Secure Bioinformatics and Genomics Cloud, Software and Services
Reliable discovery by fast BLAST of custom, standard, and SRA databases, powerful visualization, streamlined collaboration, and intuitive comparative browsing.
Researchers in hundreds of organizations trust SequenceServer; we've been cited ~300 times.
Built-in genomics & bioinformatics expertise
- AI-assisted user interface
- Intuitive visualizations
- DNA/RNA assembly, genotyping, synteny, GWAS...
No servers or UNIX necessary
- Secure, reliable & accurate
- On-demand scaling
- BLAST, SRA BLAST, genome browsers, custom visualization
Designed for collaboration among peers
- Centralized data store
- Analysis sharing & annotation
- History & audit trails
- Training & education
Genomics is key to understanding diseases, designing drugs, ensuring food quality and security, measuring and protecting the environment. It underpins bold and impactful discoveries.
SequenceServer is the best tool for BLASTing standard and custom genomes and transcriptomes, and the Sequence Read Archive (SRA). No queues or unreasonable limitations. It's also provides gene annotations, gene functions, enables genome browsing, and enables sharing and annotating data and results with peers.
Commercial and non-profit organizations worldwide use SequenceServer
Academics cite our 2019 MBE paper almost weekly (check Google Scholar or a slightly dated list here).
-
Zhiquan Yang et al.
(2024)
Nucleic Acids Research
52
SoyMD: a platform combining multi-omics data with various tools for soybean research and breeding. -
Rasmus O Jensen et al.
(2024)
Scientific Data
11
Phylogenomics and genetic analysis of solvent-producing Clostridium species. -
Jorge HernandezGarcia et al.
(2024)
Proceedings Of The National Academy Of Sciences
121
DELLA proteins recruit the Mediator complex subunit MED15 to coactivate transcription in land plants. -
Qiang He et al.
(2024)
Molecular Plant
17
A complete reference genome assembly for foxtail millet and Setaria-db, a comprehensive database for Setaria. -
Metin G Davutoglu et al.
(2024)
Communications Biology
7
Gliding motility of the diatom Craspedostauros australis coincides with the intracellular movement of raphid-specific myosins. -
Philipp Olt et al.
(2024)
Plant, Cell & Environment
47
The LaCLE35 peptide modifies rootlet density and length in cluster roots of white lupin. -
Arjen E vant Hof et al.
(2024)
Science Advances
10
Zygosity-based sex determination in a butterfly drives hypervariability of Masculinizer. -
Dongxu Liu et al.
(2024)
Nucleic Acids Research
AMIR: a multi-omics data platform for Asteraceae plants genetics and breeding research. -
Eva Pyrihova et al.
(2024)
Elife
13
A mitochondrial carrier transports glycolytic intermediates to link cytosolic and mitochondrial glycolysis in the human gut parasite Blastocystis. -
Alexander Sullivan et al.
(2024)
Nucleic Acids Research
20 years of the Bio-Analytic Resource for Plant Biology. -
Hiroto Tadano et al.
(2024)
Scientific Reports
14
Unique spatially and temporary-regulated/sex-specific expression of a long ncRNA, Nb-1, suggesting its pleiotropic functions associated with honey bee lifecycle. -
Pansee Gamaleldin et al.
(2024)
Bmc Genomics
25
Comparison of genotypic features between two groups of antibiotic resistant Klebsiella pneumoniae clinical isolates obtained before and after the COVID-19 pandemic from Egypt. -
Sen Wang et al.
(2024)
Horticulture Research
11
HortGenome Search Engine, a universal genomic search engine for horticultural crops. -
Davinder Singh et al.
(2024)
Frontiers In Plant Science
15
Genetic mapping of stripe rust resistance in a geographically diverse barley collection and selected biparental populations. -
Kyle M Benowitz et al.
(2024)
Genome Biology And Evolution
16
Fundamental patterns of structural evolution revealed by chromosome-length genomes of cactophilic Drosophila. -
Fanelie Bachelet et al.
(2024)
The Plant Journal
119
The vacuolar sulfate transporter PsSULTR4 is a key determinant of seed yield and protein composition in pea. -
Seung Chul Shin et al.
(2024)
Bmc Genomics
25
Gene loss in Antarctic icefish: evolutionary adaptations mimicking Fanconi Anemia?. -
Mana Sato et al.
(2024)
Dna Research
31
Chromosomal DNA sequences of the Pacific saury genome: versatile resources for fishery science and comparative biology. -
Daniela Polic et al.
(2024)
Molecular Ecology
33
Genetic structure, UV-vision, wing coloration and size coincide with colour polymorphism in Fabriciana adippe butterflies. -
Cao Hengchun et al.
(2024)
Database
2024
SesamumGDB: a comprehensive platform for Sesamum genetics and genomics analysis. -
John Gray et al.
(2024)
Current Plant Biology
40
GRASSIUS 2.0: A gene regulatory information knowledgebase for maize and other grasses. -
Lukasz F Sobala et al.
(2024)
Genome Biology And Evolution
16
LukProt: A database of eukaryotic predicted proteins designed for investigations of animal origins. -
Jiawei Li et al.
(2024)
Journal Of Integrative Plant Biology
SapBase: A central portal for functional and comparative genomics of Sapindaceae species. -
Xinchun Li et al.
(2024)
Marine Biotechnology
OysterDB: A Genome Database for Ostreidae. -
Joao Lozano et al.
(2024)
Microbiology Spectrum
12
Analyzing the safety of the parasiticide fungus Mucor circinelloides: first insights on its virulence profile and interactions with the avian gut microbial community. -
ChingChing Wee et al.
(2024)
Scientia Horticulturae
327
An eFP reference gene expression atlas for mangosteen. -
Jumpei Ikenaga et al.
(2024)
Biomolecules
14
Identification of Six Novel Proteins Containing a ZP Module from Nemertean Species. -
Diane Mather et al.
(2024)
Plants
13
A Quantitative Trait Locus with a Major Effect on Root-Lesion Nematode Resistance in Barley. -
Hugh Wallwork et al.
(2024)
Plant Breeding
Adult-plant resistance to net form net blotch in barley (Hordeum vulgare L.): Durability and genetic control. -
Michael J Havey et al.
(2024)
Journal Of The American Society For Horticultural Science
149
Genetic Analyses of the Shape and Volume of Onion Bulbs and Daylength Effects on Bulbing. -
Jacinta Agius et al.
(2024)
Comparative Immunology Reports
6
Analysis of the presence of anti-viral innate immune pathways in the Australian Haliotis laevigata. -
Julian M Hahnfeld et al.
(2024)
Biorxiv
sORFdb--A database for sORFs, small proteins, and small protein families in bacteria. -
Hang Yang et al.
(2024)
Biorxiv
From Understudied to Understood: Multi-Omics Analysis with MiniENCODE Exemplified by Zebrafish. -
Andi V Barker et al.
(2024)
Biorxiv
Holostean genomes reveal evolutionary novelty in the vertebrate immunoproteasome that have implications for MHCI function. -
Logan Hallee et al.
(2024)
Biorxiv
Annotation Vocabulary (Might Be) All You Need. -
Guohao Han et al.
(2024)
Biorxiv
PIGOME: An Integrated and Comprehensive Multi-omics Database for Pig Functional Genomics Studies. -
Kamawela P Leka et al.
(2024)
Methods In Enzymology
707
An introduction to comparative genomics, EukProt, and the reciprocal best hit (RBH) method for bench biologists: Ancestral phosphorylation of Tom22 in eukaryotes as a case study. -
Kevin Arthur McQuirk et al.
(2024)
Na
Rewilding shows differential fitness of Physella acuta snail populations with different invasive potential. -
Nahomy Alexandra Teran Salazar et al.
(2024)
Na
Aislamiento y an{\'a}lisis del c{\'o}digo de barras moleculares 16s de bacterias pat{\'o}genas asociadas a las enfermedades del camar{\'o}n Litopenaeus vannamei.. -
Carol J Bult et al.
(2023)
Mammalian Genome
34
The alliance of genome resources: transforming comparative genomics. -
Yuhua Fu et al.
(2023)
Nucleic Acids Research
51
IAnimal: a cross-species omics knowledgebase for animals. -
Robert Eric Hoffie et al.
(2023)
Plant Biotechnology Journal
21
Novel resistance to the Bymovirus BaMMV established by targeted mutagenesis of the PDIL5-1 susceptibility gene in barley. -
Alfia Khairullina et al.
(2023)
Plants
12
Biocontrol effect of Clonostachys rosea on Fusarium graminearum infection and mycotoxin detoxification in oat (Avena sativa). -
Biyuan Li et al.
(2023)
Frontiers In Plant Science
14
A BrLINE1-RUP insertion in BrCER2 alters cuticular wax biosynthesis in Chinese cabbage (Brassica rapa L. ssp. pekinensis). -
Rizky Pasthika Kirana et al.
(2023)
Theoretical And Applied Genetics
136
Pyramiding Fusarium head blight resistance QTL from T. aestivum, T. dicoccum and T. dicoccoides in durum wheat. -
Shrikant Sharma et al.
(2023)
Frontiers In Plant Science
14
Pho1a (plastid starch phosphorylase) is duplicated and essential for normal starch granule phenotype in tubers of Solanum tuberosum L. -
Xuzhen Li et al.
(2023)
Plant Communications
4
Teabase: A comprehensive omics database of Camellia. -
Megan ES Sorensen et al.
(2023)
Current Biology
33
A novel kleptoplastidic symbiosis revealed in the marine centrohelid Meringosphaera with evidence of genetic integration. -
Jeremy B Swann et al.
(2023)
Bmc Immunology
24
A survey of the adaptive immune genes of the polka-dot batfish Ogcocephalus cubifrons. -
Deborah A Triant et al.
(2023)
Mammalian Genome
34
AgAnimalGenomes: Browsers for viewing and manually annotating farm animal genomes. -
JanNiklas Macher et al.
(2023)
Mbio
14
Single-cell genomics reveals the divergent mitochondrial genomes of Retaria (Foraminifera and Radiolaria). -
Erika L Ellison et al.
(2023)
Genetics
225
Mutator transposon insertions within maize genes often provide a novel outward reading promoter. -
Felipe Marques de Almeida et al.
(2023)
F1000Research
12
Scalable and versatile container-based pipelines for de novo genome assembly and bacterial annotation.. -
Jihan Zhao et al.
(2023)
Journal Of Advanced Research
54
The subgenome Saccharum spontaneum contributes to sugar accumulation in sugarcane as revealed by full-length transcriptomic analysis. -
Timothy R Fallon et al.
(2023)
Bmc Bioinformatics
24
transXpress: a Snakemake pipeline for streamlined de novo transcriptome assembly and annotation. -
Jingyi Sun et al.
(2023)
Journal Of Marine Science And Engineering
11
Diversity of CO2 Concentrating Mechanisms in Macroalgae Photosynthesis: A Case Study of Ulva sp.. -
Matias Guerra et al.
(2023)
Electronic Journal Of Biotechnology
61
Draft genome of Pseudomonas sp. RGM 2987 isolated from Stevia philippiana roots reveals its potential as a plant biostimulant and potentially constitutes a novel species. -
Zhi Dong et al.
(2023)
Insect Science
30
BSFbase: The comprehensive genomic resource for a natural recycler, the black soldier fly (Hermetia illucens L.). -
Skyler Deutsch et al.
(2023)
Biology
12
Evaluation of candidates for systemic analgesia and general anesthesia in the emerging model cephalopod, Euprymna berryi. -
Stefan Brott et al.
(2023)
Applied Microbiology And Biotechnology
107
Unique alcohol dehydrogenases involved in algal sugar utilization by marine bacteria. -
Alliance of Genome Resources Consortium et al.
(2023)
Biorxiv
Updates to the alliance of genome resources central infrastructure alliance of genome resources consortium. -
Junhao Zhuge et al.
(2023)
International Journal Of Molecular Sciences
24
The Plant Parasitic Nematodes Database: A Comprehensive Genomic Data Platform for Plant Parasitic Nematode Research. -
Jean Franco Castro et al.
(2023)
Plant Disease
107
First report of Colletotrichum fioriniae causing anthracnose fruit rot on Vaccinium corymbosum in Chile. -
Lukasz F Sobala et al.
(2023)
Glycobiology
33
Evolution and phylogenetic distribution of endo-$\alpha$-mannosidase. -
Parva Kumar Sharma et al.
(2023)
Database
2023
An online database for einkorn wheat to aid in gene discovery and functional genomics studies. -
Michael Schorpp et al.
(2023)
European Journal Of Immunology
53
Foxn1 is not essential for T-cell development in teleosts. -
Zhaoheng Zhang et al.
(2023)
Seed Biology
2
A comprehensive atlas of long non-coding RNAs provides insight into grain development in wheat. -
Gaoqun Zhang et al.
(2023)
European Journal Of Immunology
53
Lymphocyte pathway analysis using naturally lymphocyte-deficient fish. -
Hadi Nayebi Gavgani et al.
(2023)
Plant Gene Regulatory Networks …
Methodology for Constructing a Knowledgebase for Plant Gene Regulation Information. -
S Deutsch et al.
(2023)
Na
Evaluation of Candidates for Systemic Analgesia and General Anesthesia in the Emerging Model Cephalopod, Euprymna berryi. Biology 2023, 12, 201. -
Akshay Singh et al.
(2023)
Frontiers In Plant Science
14
Amaranth Genomic Resource Database: an integrated database resource of Amaranth genes and genomics. -
Richard S Gunasekera et al.
(2023)
Plos One
18
ORFanID: A web-based search engine for the discovery and identification of orphan and taxonomically restricted genes. -
Zohra Chaddad et al.
(2023)
Plant And Soil
493
Genetic diversity, phenotypic traits, and symbiotic efficiency of native Bradyrhizobium strains of Lupinus luteus in Morocco. -
Carolina CamposQuiroz et al.
(2023)
Taxonomy
3
Description of Two Fungal Endophytes Isolated from Fragaria chiloensis subsp. chiloensis f. patagonica: Coniochaeta fragariicola sp. nov. and a New Record of Coniochaeta hansenii. -
Yunchi Zhu et al.
(2023)
Database
2023
MineProt: a stand-alone server for structural proteome curation. -
Bernice Waweru et al.
(2023)
Biorxiv
Chromosome-scale assembly of the African yam bean genome. -
Suzanne A Aleksander et al.
(2023)
Biorxiv
Updates to the Alliance of Genome Resources Central Infrastructure Alliance of Genome Resources Consortium. -
Agnieszka P Lipinska et al.
(2023)
Genome Biology And Evolution
15
The Rhodoexplorer Platform for red algal genomics and whole-genome assemblies for several Gracilaria species. -
Erika L Magnusson et al.
(2023)
Na
Mutator transposons in Zea mays impact transcriptional regulatory networks and underlying gene expression. -
Kara Jones et al.
(2023)
Na
Placing the Evolutionary History of Desmognathus Salamanders in Context: A Phylogeographic Approach. -
Sulun Zheng et al.
(2023)
Na
Characterizing Candidate Genes in Apple Flesh Development. -
Lianguang Shang et al.
(2022)
Cell Research
32
A super pan-genomic landscape of rice. -
Haixu Chen et al.
(2022)
Nucleic Acids Research
50
BRAD V3. 0: an upgraded Brassicaceae database. -
Immo Burkhardt et al.
(2022)
Nature Chemical Biology
18
Ancient plant-like terpene biosynthesis in corals. -
Daniel J Richter et al.
(2022)
Peer Community Journal
2
EukProt: a database of genome-scale predicted proteins across the diversity of eukaryotes. -
Desnor N Chigumba et al.
(2022)
Nature Chemical Biology
18
Discovery and biosynthesis of cyclic plant peptides via autocatalytic cyclases. -
Eric Yao et al.
(2022)
Database
2022
GrainGenes: a data-rich repository for small grains genetics and genomics. -
Terry J TorresCruz et al.
(2022)
Plant Disease
106
FUSARIUM-ID v. 3.0: an updated, downloadable resource for Fusarium species identification. -
Roland D Kersten et al.
(2022)
Journal Of The American Chemical Society
144
Gene-guided discovery and ribosomal biosynthesis of moroidin peptides. -
Amy T Walsh et al.
(2022)
Nucleic Acids Research
50
Hymenoptera Genome Database: new genomes and annotation datasets for improved go enrichment and orthologue analyses. -
Hang Yan et al.
(2022)
Frontiers In Cellular And Infection Microbiology
12
Comparison of the gut microbiota in different age groups in China. -
JieDan Chen et al.
(2022)
Frontiers In Plant Science
13
TeaGVD: a comprehensive database of genomic variations for uncovering the genetic architecture of metabolic traits in tea plants. -
Jonathan C Chen et al.
(2022)
Nature Communications
13
Generating experimentally unrelated target molecule-binding highly functionalized nucleic-acid polymers using machine learning. -
Fanbo Meng et al.
(2022)
Horticulture Research
9
TCMPG: an integrative database for traditional Chinese medicine plant genomes. -
Gaetan Droc et al.
(2022)
Horticulture Research
9
The banana genome hub: a community database for genomics in the Musaceae. -
Shuhei Yabe et al.
(2022)
Isme Communications
2
Vulcanimicrobium alpinus gen. nov. sp. nov., the first cultivated representative of the candidate phylum “Eremiobacterota”, is a metabolically versatile aerobic anoxygenic phototroph. -
Aspasia Kontou et al.
(2022)
Traffic
23
Evolution of factors shaping the endoplasmic reticulum. -
Lu Gan et al.
(2022)
Proceedings Of The National Academy Of Sciences
119
Divergent evolution of extreme production of variant plant monounsaturated fatty acids. -
Zhibin Zhang et al.
(2022)
Frontiers In Plant Science
13
GRAND: an integrated genome, transcriptome resources, and gene network database for gossypium. -
Yong Wang et al.
(2022)
Frontiers In Plant Science
13
BoGDB: An integrative genomic database for Brassica oleracea L.. -
Maxime Deraspe et al.
(2022)
Scientific Reports
12
Flexible protein database based on amino acid k-mers. -
Matthew J Tuttle et al.
(2022)
Msphere
7
Plasmid-mediated stabilization of prophages. -
Shengmei Li et al.
(2022)
Genes
13
Revealing genetic differences in fiber elongation between the offspring of sea island cotton and upland cotton backcross populations based on transcriptome and weighted gene coexpression networks. -
David J Krause et al.
(2022)
Molecular Biology And Evolution
39
Functional divergence in a multi-gene family is a key evolutionary innovation for anaerobic growth in Saccharomyces cerevisiae. -
Ryoma Takeshima et al.
(2022)
Bmc Plant Biology
22
Genetic basis of maturity time is independent from that of flowering time and contributes to ecotype differentiation in common buckwheat (Fagopyrum esculentum Moench). -
Jingwei Song et al.
(2022)
Marine Biotechnology
24
Comparative transcriptomics of the northern quahog Mercenaria mercenaria and southern quahog Mercenaria campechiensis in response to chronic heat stress. -
Ran Tian et al.
(2022)
Frontiers In Ecology And Evolution
10
Molecular evolution of vision-related genes may contribute to marsupial photic niche adaptations. -
Sven Redsun et al.
(2022)
… : Methods And Protocols
Doing genetic and genomic biology using the legume information system and associated resources. -
Michael Niederwanger et al.
(2022)
Frontiers In Environmental Science
10
Expression of biomarkers connected to endocrine disruption in Cottus gobio and Salmo trutta fario in relation to sewage treatment plant-efflux and pesticides. -
Kara B Carlson et al.
(2022)
Conservation Genetics
23
Transcriptome annotation reveals minimal immunogenetic diversity among Wyoming toads, Anaxyrus baxteri. -
Yunchi Zhu et al.
(2022)
Gigascience
11
Utilizing an artificial intelligence system to build the digital structural proteome of reef-building corals. -
Iman Almansour et al.
(2022)
Database
2022
hCoronavirusesDB: an integrated bioinformatics resource for human coronaviruses. -
Nobuko Sumiya et al.
(2022)
Protoplasma
259
Coordination mechanism of cell and cyanelle division in the glaucophyte alga Cyanophora sudae. -
Kyle M Benowitz et al.
(2022)
Biorxiv
Chromosome-length genome assemblies of cactophilic Drosophila illuminate links between structural and sequence evolution. -
Victoria C Blake et al.
(2022)
Foods
11
GrainGenes: tools and content to assist breeders improving oat quality. -
Philip Bertemes et al.
(2022)
Marine Drugs
20
Sticking together an updated model for temporary adhesion. -
Wenlei Guo et al.
(2022)
Frontiers In Plant Science
13
GROP: A genomic information repository for oilplants. -
Domitille Jarrige et al.
(2022)
G3
12
High-quality genome of the basidiomycete yeast Dioszegia hungarica PDD-24b-2 isolated from cloud water. -
Kathryn C Lesneski et al.
(2022)
Biorxiv
Holobiont transcriptomes for the critically endangered staghorn coral (Acropora cervicornis) from two environmentally distinct sites on Turneffe Atoll, Belize. -
Yunchi Zhu et al.
(2022)
Arxiv Preprint Arxiv:2212.07809
MineProt: modern application for custom protein curation. -
HuiSu Kim et al.
(2022)
Gigabyte
2022
LT1, an ONT long-read-based assembly scaffolded with Hi-C data and polished with short reads. -
Konstantinos Sousounis et al.
(2022)
Salamanders: Methods And …
A Practical Guide for CRISPR-Cas9-Induced Mutations in Axolotls. -
Daniel J Richter et al.
(2022)
Na
Peer Community Journal. -
Varnika Mittal et al.
(2022)
Bmc Genomic Data
23
EchinoDB: an update to the web-based application for genomic and transcriptomic data on echinoderms. -
Jean Franco Castro et al.
(2022)
Microbiology Resource Announcements
11
Draft Genome Sequence of Pseudomonas sp. Strain RGM 3321, a Phyllosphere Endophyte from Fragaria chiloensis subsp. chiloensis f. patagonica. -
Charikleia Karageorgiou et al.
(2022)
Na
Development of a high-quality annotated reference genome and evolutionary genomics analysis of chromosomal inversions in Drosophila subobscura. -
Lakshmi Sahitya Appikatla et al.
(2022)
Na
Identifying Carotenoid-Associated Transcription Factors in Yellow-fleshed Apples. -
VC Blake et al.
(2022)
Na
GrainGenes: Tools and Content to Assist Breeders Improving Oat Quality. Foods 2022, 11, 914. -
櫻岡良平 et al.
(2022)
Na
比較ゲノム解析に基づく細菌の抗菌物質生産機構の多様性に関する研究. -
Joseph J Braymer et al.
(2021)
Biochimica Et Biophysica Acta (Bba)-Molecular Cell Research
1868
Mechanistic concepts of iron-sulfur protein biogenesis in Biology. -
Max E Schon et al.
(2021)
Nature Communications
12
Single cell genomics reveals plastid-lacking Picozoa are close relatives of red algae. -
Laurence Garczarek et al.
(2021)
Nucleic Acids Research
49
Cyanorak v2. 1: a scalable information system dedicated to the visualization and expert curation of marine and brackish picocyanobacteria genomes. -
Tim Koopmans et al.
(2021)
Npj Regenerative Medicine
6
Ischemic tolerance and cardiac repair in the spiny mouse (Acomys). -
Robert Eric Hoffie et al.
(2021)
Frontiers In Genome Editing
3
Targeted knockout of eukaryotic translation initiation factor 4E confers bymovirus resistance in winter barley. -
Kathy Darragh et al.
(2021)
Plos Biology
19
A novel terpene synthase controls differences in anti-aphrodisiac pheromone production between closely related Heliconius butterflies. -
Ana B Tinoco et al.
(2021)
Elife
10
Ancient role of sulfakinin/cholecystokinin-type signalling in inhibitory regulation of feeding processes revealed in an echinoderm. -
Carlos Caurcel et al.
(2021)
Philosophical Transactions Of The Royal Society B
376
MolluscDB: a genome and transcriptome database for molluscs. -
Satoshi Miyazaki et al.
(2021)
Scientific Reports
11
Evolutionary transition of doublesex regulation from sex-specific splicing to male-specific transcription in termites. -
Xiaogang Lei et al.
(2021)
Beverage Plant Research
1
TeaPGDB: tea plant genome database. -
Jie Zhou et al.
(2021)
Frontiers In Genetics
12
Genome-wide identification and expression analysis of the plant U-box protein gene family in Phyllostachys edulis. -
Tullia I Terraneo et al.
(2021)
Molecular Phylogenetics And Evolution
161
Phylogenomics of Porites from the Arabian Peninsula. -
Ilya Kirov et al.
(2021)
Plants
10
Transposons hidden in Arabidopsis thaliana genome assembly gaps and mobilization of non-autonomous LTR retrotransposons unravelled by nanotei pipeline. -
Thomas M Adams et al.
(2021)
Bmc Genomics
22
Rust expression browser: an open source database for simultaneous analysis of host and pathogen gene expression profiles with expVIP. -
Cyprien Guerin et al.
(2021)
Isme Communications
1
Transcriptome architecture and regulation at environmental transitions in flavobacteria: the case of an important fish pathogen. -
Linglong Zhu et al.
(2021)
Crop Science
61
High-density linkage map construction and QTL analysis of fiber quality and lint percentage in tetraploid cotton. -
Yuhan Zhou et al.
(2021)
Horticulturae
8
GDS: a genomic database for strawberries (Fragaria spp.). -
Philip Bertemes et al.
(2021)
International Journal Of Molecular Sciences
22
(Un) expected similarity of the temporary adhesive systems of marine, brackish, and freshwater flatworms. -
Sofia N Barreira et al.
(2021)
Molecular Biology And Evolution
38
Aniprotdb: a collection of consistently generated metazoan proteomes for comparative genomics studies. -
Giovana Acha et al.
(2021)
Plants
10
A traceable DNA-Replicon derived vector to speed up gene editing in potato: interrupting genes related to undesirable postharvest tuber traits as an example. -
Clive H Bock et al.
(2021)
Phytopathology{Textregistered}
111
Mating type idiomorphs, heterothallism, and high genetic diversity in Venturia carpophila, cause of peach scab. -
Tomohiro Morohoshi et al.
(2021)
Bioscience, Biotechnology, And Biochemistry
85
Comparative genome analysis reveals the presence of multiple quorum-sensing systems in plant pathogenic bacterium, Erwinia rhapontici. -
Dahui Hu et al.
(2021)
Rna Bioinformatics
Database resources for functional circular RNAs. -
Max E Schon et al.
(2021)
Biorxiv
Picozoa are archaeplastids without plastid. -
Chi Zhang et al.
(2021)
Gene
772
NBIGV-DB: A dedicated database of non-B cell derived immunoglobulin variable region. -
Huiyuan Wang et al.
(2021)
Frontiers In Plant Science
12
PSDX: A comprehensive multi-omics association database of Populus trichocarpa with a focus on the secondary growth in response to stresses. -
Berenice TalamantesBecerra et al.
(2021)
Softwarex
14
omicR: A tool to facilitate BLASTn alignments for sequence data. -
Ryohei Sakuraoka et al.
(2021)
Microbes And Environments
36
Comparative Genome Analysis Reveals Differences in Biocontrol Efficacy According to Each Individual Isolate Belonging to Rhizospheric Fluorescent Pseudomonads. -
Hyungtaek Jung et al.
(2021)
Biorxiv
easyfm: An easy software suite for file manipulation of Next Generation Sequencing data on desktops. -
Satoshi Miyazaki et al.
(2021)
Biorxiv
Evolutionary transition of doublesex regulation in termites and cockroaches: from sex-specific splicing to male-specific transcription. -
Sreedevi Padmanabhan et al.
(2021)
Micropublication Biology
2021
Identification and in silico analysis of the origin recognition complex in the human fungal pathogen Candida albicans. -
Aaron DeLeo et al.
(2021)
Na
A Phylogenetic Study of the Duplication of (6-4) Photolyase DNA Repair Enzymes within Plant Pathogen Genus Phytophthora. -
Sara Calatayud Robert et al.
(2021)
Na
Modular evolution of domain repeat proteins. Metal-binding and domain repeats of Metallothioneins in molluscs and chordates. -
Tereza vCalounova et al.
(2021)
Na
De novo transcriptomics and its use in non-model organisms. -
Louise N Perez et al.
(2021)
The International Journal Of Developmental Biology
65
The subterranean catfish Phreatobius cisternarum provides insights into visual adaptations to the phreatic environment. -
HuiSu Kim et al.
(2021)
Biorxiv
The Lithuanian reference genome LT1-a human de novo genome assembly with short and long read sequence and Hi-C data. -
Michele Cosi et al.
(2021)
Journal Of Open Source Education
4
StarBLAST: a scalable BLAST+ solution for the classroom. -
Juliette Rousseaux et al.
(2021)
Na
Bioluminescence chez Amphiura filiformis (OF M{\"u}ller): histoire {\'e}volutive et expression de la lucif{\'e}rase. -
Jeremy B Swann et al.
(2020)
Science
369
The immunogenetics of sexual parasitism. -
Tomas Alves et al.
(2020)
Personality And Individual Differences
155
Incorporating personality in user interface design: A review. -
Songtao Gui et al.
(2020)
Iscience
23
ZEAMAP, a comprehensive database adapted to the maize multi-omics era. -
Yuhua Fu et al.
(2020)
Communications Biology
3
A gene prioritization method based on a swine multi-omics knowledgebase and a deep learning model. -
Colin Y Kim et al.
(2020)
Nature Communications
11
The chloroalkaloid (-)-acutumine is biosynthesized via a Fe (II)-and 2-oxoglutarate-dependent halogenase in Menispermaceae plants. -
Hanna Koch et al.
(2020)
Scientific Reports
10
Genomic, metabolic and phenotypic variability shapes ecological differentiation and intraspecies interactions of Alteromonas macleodii. -
Ya Zhang et al.
(2020)
Open Biology
10
Molecular and functional characterization of somatostatin-type signalling in a deuterostome invertebrate. -
Dehe Wang et al.
(2020)
Nucleic Acids Research
48
MaGenDB: a functional genomics hub for Malvaceae plants. -
Yingshu Li et al.
(2020)
Genome Research
30
Dynamic transcriptional and chromatin accessibility landscape of medaka embryogenesis. -
Luis Alfonso YanezGuerra et al.
(2020)
Elife
9
Echinoderms provide missing link in the evolution of PrRP/sNPF-type neuropeptide signalling. -
Md Shamimuzzaman et al.
(2020)
Nucleic Acids Research
48
Bovine Genome Database: new annotation tools for a new reference genome. -
Wenyi Wu et al.
(2020)
Database
2020
PncStress: a manually curated database of experimentally validated stress-responsive non-coding RNAs in plants. -
Liying Yu et al.
(2020)
Database
2020
SAGER: a database of Symbiodiniaceae and Algal Genomic Resource. -
Nathan S Hart et al.
(2020)
Molecular Biology And Evolution
37
Visual opsin diversity in sharks and rays. -
Reinhard Dallinger et al.
(2020)
Metallomics
12
Metallomics reveals a persisting impact of cadmium on the evolution of metal-selective snail metallothioneins. -
Benedicte Billard et al.
(2020)
Current Biology
30
A natural mutational event uncovers a life history trade-off via hormonal pleiotropy. -
R Travis Moreland et al.
(2020)
Database
2020
The Mnemiopsis Genome Project Portal: integrating new gene expression resources and improving data visualization. -
SJ Reichler et al.
(2020)
Journal Of Dairy Science
103
Interventions designed to control postpasteurization contamination in high-temperature, short-time-pasteurized fluid milk processing facilities: A case study on the effect of employee training, clean-in-place chemical modification, and preventive maintenance programs. -
Jeremy Gauthier et al.
(2020)
Molecular Ecology
29
Contrasting genomic and phenotypic outcomes of hybridization between pairs of mimetic butterfly taxa across a suture zone. -
Jessica A Goodheart et al.
(2020)
Scientific Reports
10
Laboratory culture of the California Sea Firefly Vargula tsujii (Ostracoda: Cypridinidae): Developing a model system for the evolution of marine bioluminescence. -
Robert M Waterhouse et al.
(2020)
Immunity In Insects
Characterization of insect immune systems from genomic data. -
Jiongliang Wang et al.
(2020)
Frontiers In Genetics
11
Coexpression analysis reveals dynamic modules regulating the growth and development of cirri in the rattans (Calamus simplicifolius and Daemonorops jenkinsiana). -
DA Triant et al.
(2020)
Animal Genetics
51
Using online tools at the Bovine Genome Database to manually annotate genes in the new reference genome. -
Ana B Tinoco et al.
(2020)
Biorxiv
Evolutionarily ancient role of cholecystokinin-type neuropeptide signalling as an inhibitory regulator of feeding-related processes revealed in an echinoderm. -
Karam B Singh et al.
(2020)
The Lupin Genome
Overview of genomic resources available for lupins with a focus on narrow-leafed lupin (Lupinus angustifolius). -
Abdelkader Ainouche et al.
(2020)
The Lupin Genome
The repetitive content in lupin genomes. -
Sofia N Barreira et al.
(2020)
Biorxiv
AniProtDB: A Collection of Uniformly Generated Metazoan Proteomes for Comparative Genomics Studies. -
Karam B Singh et al.
(2020)
The Lupin Genome
and Lars G. Kamphuis. -
Meinhard Simon et al.
(2020)
Na
Genomic, metabolic and phenotypic variability shapes ecological differentiation and intraspecies interactions of Alteromonas macleodii. -
Armel Salmon et al.
(2020)
The Lupin Genome
Abdelkader A{\"\i}nouche, Aurore Paris, Delphine Giraud, Jean Keller, Pauline Raimondeau, Fr{\'e}d{\'e}ric Mah{\'e}, Pavel Neuman, Petr Novak, Jiri Macas, Malika A{\"\i}nouche. -
Milot Mirdita et al.
(2019)
Bioinformatics
35
MMseqs2 desktop and local web server app for fast, interactive sequence searches. -
DeAnna C Bublitz et al.
(2019)
Cell
179
Peptidoglycan production by an insect-bacterial mosaic. -
Xianwen Meng et al.
(2019)
Database
2019
CircFunBase: a database for functional circular RNAs. -
Tomavs Pluskal et al.
(2019)
Nature Plants
5
The biosynthetic origin of psychoactive kavalactones in kava. -
Olesya Levsh et al.
(2019)
Journal Of Biological Chemistry
294
Independent evolution of rosmarinic acid biosynthesis in two sister families under the Lamiids clade of flowering plants. -
Vera Thole et al.
(2019)
Bmc Genomics
20
RNA-seq, de novo transcriptome assembly and flavonoid gene analysis in 13 wild and cultivated berry fruit species with high content of phenolics. -
Muyan Chen et al.
(2019)
Scientific Reports
9
Neuropeptide precursors and neuropeptides in the sea cucumber Apostichopus japonicus: a genomic, transcriptomic and proteomic analysis. -
Natasha A Botwright et al.
(2019)
G3: Genes, Genomes, Genetics
9
Greenlip abalone (Haliotis laevigata) genome and protein analysis provides insights into maturation and spawning. -
Tepsuda Rungrat et al.
(2019)
Plant Direct
3
A Genome-Wide Association Study of Non-Photochemical Quenching in response to local seasonal climates in Arabidopsis thaliana. -
YuLong Li et al.
(2019)
Genome Biology And Evolution
11
Population genomic signatures of genetic structure and environmental selection in the catadromous roughskin sculpin Trachidermus fasciatus. -
Suda Parimala Ravindran et al.
(2019)
Scientific Reports
9
Daphnia s tressor database: Taking advantage of a decade of Daphnia ‘-omics’ data for gene annotation. -
Marcin Nowicki et al.
(2019)
Scientific Reports
9
Taraxacum kok-saghyz (rubber dandelion) genomic microsatellite loci reveal modest genetic diversity and cross-amplify broadly to related species. -
Allan M CarrilloBaltodano et al.
(2019)
Journal Of Morphology
280
Developmental architecture of the nervous system in Themiste lageniformis (Sipuncula): new evidence from confocal laser scanning microscopy and gene expression. -
Iman Almansour et al.
(2019)
Infection, Genetics And Evolution
75
MMRdb: Measles, mumps, and rubella viruses database and analysis resource. -
Y Liang et al.
(2019)
Insect Molecular Biology
28
Developmental expression and evolution of hexamerin and haemocyanin from Folsomia candida (Collembola). -
MengYu Li et al.
(2019)
Mitochondrial Dna Part B
4
First mitochondrial genome of a periwinkle from the genus Littoraria: Littoraria sinensis. -
Kathy Darragh et al.
(2019)
Biorxiv
A novel terpene synthase produces an anti-aphrodisiac pheromone in the butterfly Heliconius melpomene. -
Melissa Bizzarri et al.
(2019)
Frontiers In Genetics
10
Interplay of chimeric mating-type loci impairs fertility rescue and accounts for intra-strain variability in Zygosaccharomyces rouxii interspecies hybrid ATCC42981. -
David Aciole Barbosa et al.
(2019)
Plos Neglected Tropical Diseases
13
ParaDB: A manually curated database containing genomic annotation for the human pathogenic fungi Paracoccidioides spp.. -
Humberto Prieto et al.
(2019)
The Grape Genome
Grape biotechnology: past, present, and future. -
Luis Alfonso YanezGuerra et al.
(2019)
Biorxiv
Urbilaterian origin and evolution of sNPF-type neuropeptide signalling. -
Michael Elsy et al.
(2019)
International Journal Of Developmental Biology
63
Xenopus laevis FGF16 activates the expression of genes coding for the transcription factors Sp5 and Sp5l. -
Geoffrey Liou et al.
(2019)
Na
Enzyme structure, function, and evolution in flavonoid biosynthesis. -
Ismail Moghul et al.
(2019)
Methods Mol Biol
Correction to: Choosing the Best Gene Predictions with GeneValidator.. -
Ismail Moghul et al.
(2019)
Gene Prediction: Methods And Protocols
Choosing the Best Gene Predictions with GeneValidator. -
Evelyn Sanchez et al.
(2019)
The Grape Genome
Humberto Prieto, Mar{\'\i}a Miccono, Carlos Aguirre. -
Els LISMONT et al.
(2019)
Na
USING BRET-BASED G PROTEIN BIOSENSORS TO UNRAVEL THE G PROTEIN MEDIATED PATHWAYS IN INSECTS. -
Melissa Bizzarri et al.
(2019)
Na
Studio dei meccanismi molecolari determinanti l’identit{\`a} cellulare e la sterilit{\`a} in un ibrido allodiploide Zygosaccharomyces attraverso la biologia sintetica e l’assemblaggio de novo del genoma con le piattaforme MiSeq e MinION. -
Timothy R Fallon et al.
(2018)
Elife
7
Firefly genomes illuminate parallel origins of bioluminescence in beetles. -
Michael P TorrensSpence et al.
(2018)
Molecular Plant
11
Complete pathway elucidation and heterologous reconstitution of Rhodiola salidroside biosynthesis. -
Roland D Kersten et al.
(2018)
Proceedings Of The National Academy Of Sciences
115
Gene-guided discovery and engineering of branched cyclic peptides in plants. -
SJ Reichler et al.
(2018)
Journal Of Dairy Science
101
Pseudomonas fluorescens group bacterial strains are responsible for repeat and sporadic postpasteurization contamination and reduced fluid milk shelf life. -
Saowaros SuwansaArd et al.
(2018)
Peptides
99
Transcriptomic discovery and comparative analysis of neuropeptide precursors in sea cucumbers (Holothuroidea). -
Allah Ditta et al.
(2018)
International Journal Of Molecular Sciences
19
Assessment of genetic diversity, population structure, and evolutionary relationship of uncharacterized genes in a novel germplasm collection of diploid and allotetraploid Gossypium accessions using EST and genomic SSR markers. -
Simon Blanchoud et al.
(2018)
Scientific Reports
8
De novo draft assembly of the Botrylloides leachii genome provides further insight into tunicate evolution. -
James S Borrell et al.
(2018)
Heredity
121
Genetic diversity maintained among fragmented populations of a tree undergoing range contraction. -
Alison W Roberts et al.
(2018)
The Plant Cell
30
Functional characterization of a glycosyltransferase from the moss Physcomitrella patens involved in the biosynthesis of a novel cell wall arabinoglucan. -
Michael Patrick TorrensSpence et al.
(2018)
Acs Chemical Biology
13
Monoamine biosynthesis via a noncanonical calcium-activatable aromatic amino acid decarboxylase in psilocybin mushroom. -
Stephen J Holland et al.
(2018)
Proceedings Of The National Academy Of Sciences
115
Expansions, diversification, and interindividual copy number variations of AID/APOBEC family cytidine deaminase genes in lampreys. -
Martin Dvorak et al.
(2018)
Metallomics
10
Metal binding functions of metallothioneins in the slug Arion vulgaris differ from metal-specific isoforms of terrestrial snails. -
Blake E Sanders et al.
(2018)
Msphere
3
FusoPortal: an interactive repository of hybrid MinION-sequenced Fusobacterium genomes improves gene identification and characterization. -
Nicola Bordin et al.
(2018)
Database
2018
PVCbase: an integrated web resource for the PVC bacterial proteomes. -
Carolyn A Young et al.
(2018)
Phytopathology
108
Evidence for sexual reproduction: identification, frequency, and spatial distribution of Venturia effusa (pecan scab) mating type idiomorphs. -
Christine G Elsik et al.
(2018)
Eukaryotic Genomic Databases: Methods And Protocols
Hymenoptera genome database: using hymenopteramine to enhance genomic studies of hymenopteran insects. -
Parice A Brandies et al.
(2018)
Peerj
6
Disentangling the mechanisms of mate choice in a captive koala population. -
Trevor D Lamb et al.
(2018)
Open Biology
8
Evolution of the shut-off steps of vertebrate phototransduction. -
Els Lismont et al.
(2018)
General And Comparative Endocrinology
258
Molecular cloning and characterization of the SIFamide precursor and receptor in a hymenopteran insect, Bombus terrestris. -
Darren E Hagen et al.
(2018)
Eukaryotic Genomic Databases: Methods And Protocols
Bovine Genome Database: Tools for mining the Bos taurus genome. -
M Tangherlini et al.
(2018)
Bmc Bioinformatics
19
GLOSSary: the GLobal Ocean 16S subunit web accessible resource. -
Junhao Chen et al.
(2018)
Database
2018
MGH: a genome hub for the medicinal plant maca (Lepidium meyenii). -
Jared Mamrot et al.
(2018)
Biorxiv
Embryonic gene transcription in the spiny mouse (Acomys cahirinus): an investigtion into the embryonic genome activation. -
Tomavs Pluskal et al.
(2018)
Biorxiv
The biosynthetic origin of psychoactive kavalactones in kava. -
Suda Parimala Ravindran et al.
(2018)
Na
Comparative and population transcriptomics of Daphnia galeata. -
Turkan Bengu Tacsdan et al.
(2018)
Na
Ay{\c{c}}i{\c{c}}e{\u{g}}i (Helianthus annuus L.) bitkisinde b{\"u}y{\"u}me d{\"u}zenleyici fakt{\"o}r (GRF) transkripsiyon fakt{\"o}r{\"u} gen ailesinin genom boyutunda in silico tan{\i}mlanmas{\i} ve karakterizasyonu. -
Thierry Naas et al.
(2017)
Journal Of Enzyme Inhibition And Medicinal Chemistry
32
Beta-lactamase database (BLDB)--structure and function. -
James K Hane et al.
(2017)
Plant Biotechnology Journal
15
A comprehensive draft genome sequence for lupin (Lupinus angustifolius), an emerging health food: insights into plant--microbe interactions and legume evolution. -
Tao Zhu et al.
(2017)
Bmc Plant Biology
17
CottonFGD: an integrated functional genomics database for cotton. -
YaTing Chao et al.
(2017)
Plant And Cell Physiology
58
Orchidstra 2.0—A transcriptomics resource for the orchid family. -
Domonique A Carson et al.
(2017)
Applied And Environmental Microbiology
83
Bacteriocins of non-aureus staphylococci isolated from bovine milk. -
Rodrigo Pracana et al.
(2017)
Evolution Letters
1
Fire ant social chromosomes: Differences in number, sequence and expression of odorant binding proteins. -
Steven T Hill et al.
(2017)
Database
2017
HopBase: a unified resource for Humulus genomics. -
Jay Shockey et al.
(2017)
Planta
245
Naturally occurring high oleic acid cottonseed oil: identification and functional analysis of a mutant allele of Gossypium barbadense fatty acid desaturase-2. -
Felipe Torres et al.
(2017)
Database
2017
LeishDB: a database of coding gene annotation and non-coding RNAs in Leishmania braziliensis. -
Benjamin J Hunt et al.
(2017)
Ecology And Evolution
7
The Euphausia superba transcriptome database, Superba SE: An online, open resource for researchers. -
Richard J Challis et al.
(2017)
Database
2017
GenomeHubs: simple containerized setup of a custom Ensembl database and web server for any species. -
James Borrell et al.
(2017)
Na
Ecological Genomics for the Conservation of Dwarf Birch. -
XingXing Shen et al.
(2016)
G3: Genes, Genomes, Genetics
6
Reconstructing the backbone of the Saccharomycotina yeast phylogeny using genome-scale data. -
Thomas R Connor et al.
(2016)
Microbial Genomics
2
CLIMB (the Cloud Infrastructure for Microbial Bioinformatics): an online resource for the medical microbiology community. -
Terry Mun et al.
(2016)
Scientific Reports
6
Lotus Base: An integrated information portal for the model legume Lotus japonicus. -
Yi Jin Liew et al.
(2016)
Database
2016
Reefgenomics. Org-a repository for marine genomics data. -
Ryan F McCormick et al.
(2016)
Plant Physiology
172
3D sorghum reconstructions from depth images identify QTL regulating shoot architecture. -
Richard J Challi et al.
(2016)
Biorxiv
Lepbase: the Lepidopteran genome database. -
Dean C Semmens et al.
(2016)
Open Biology
6
Transcriptomic identification of starfish neuropeptide precursors yields new insights into neuropeptide evolution. -
So Nakagawa et al.
(2016)
Database
2016
gEVE: a genome-based endogenous viral element database provides comprehensive viral protein-coding sequences in mammalian genomes. -
Daniel A Janies et al.
(2016)
Bmc Bioinformatics
17
EchinoDB, an application for comparative transcriptomics of deeply-sampled clades of echinoderms. -
MP TorrensSpence et al.
(2016)
Methods In Enzymology
576
A workflow for studying specialized metabolism in nonmodel eukaryotic organisms. -
Steven T Hill et al.
(2016)
Na
The pursuit of hoppiness: propelling hop into the genomic era. -
Bruno Louro et al.
(2016)
Marine Genomics
30
Having a BLAST: Searchable transcriptome resources for the gilthead sea bream and the European sea bass. -
Benjamin James Hunt et al.
(2016)
Na
Advancing molecular crustacean chronobiology through the characterisation of the circadian clock in two malacostracan species, Euphausia superba and Parhyale hawaiensis. -
Martin Page et al.
(2016)
Bmc Research Notes
9
blastjs: a BLAST+ wrapper for Node. js. -
Richard J Challis et al.
(2016)
Peerj Preprints
4
EasyMirror and EasyImport: Simplifying the setup of a custom Ensembl database and webserver for any species. -
Ryan F McCormick et al.
(2016)
Biorxiv
3D sorghum reconstructions from depth images enable identification of quantitative trait loci regulating shoot architecture. -
Ioannis Kirmitzoglou et al.
(2015)
Bioinformatics
31
LCR-eXXXplorer: a web platform to search, visualize and share data for low complexity regions in protein sequences. -
Parichit Sharma et al.
(2014)
Plos One
9
WImpiBLAST: Web interface for mpiBLAST to help biologists perform large-scale annotation using high performance computing. -
Ioannis K Kirmitzoglou et al.
(2014)
Na
Development of algorithms and software for unravelling the biological role of low complexity regions in protein sequences. -
Luis Carlos Belarmino et al.
(2013)
Bmc Bioinformatics
14
SymGRASS: a database of sugarcane orthologous genes involved in arbuscular mycorrhiza and root nodule symbiosis.
Benefits of using SequenceServer Cloud
Just do the research
Focus on the science, not UNIX, servers, updates or waiting for NCBI.
Simply upload FASTA files or select standard sequence databases.
Begin BLASTing within seconds.
~300 citations can't be wrong.
Focus on the biology.
Powerful visualizations
Gain deep insight through diverse visualization approaches.
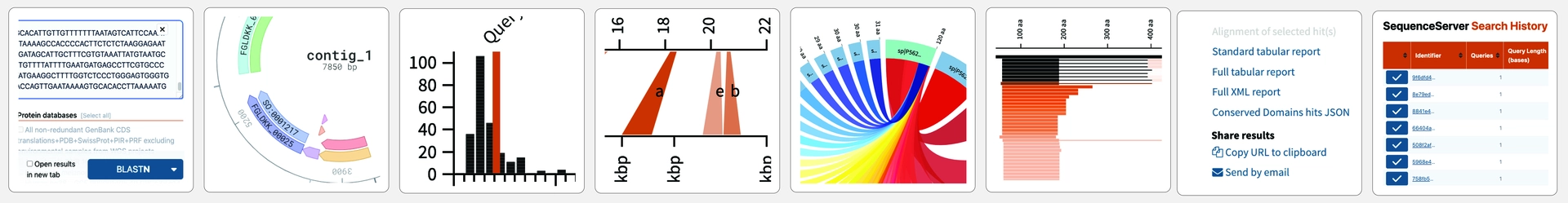
All BLAST results include pairwise alignment graphs, Circos-style overview plots, and length distribution histograms. These plots clarify and help to interpret analysis results. Publication-ready export in .PNG or .SVG formats. You work faster and more accurately.
Intelligent user interface
Faster & more accurate.
Our user interface elegantly aims to make you happy. "AI" detection of nucleotides vs amino-acids. Automagic FASTQ to FASTA conversion. Automagic selection of the best BLAST method.
Such features accelerate your workflow and reduce your chances of making errors.
Share and collaborate
Have a central SequenceServer database for your team.
Share databases and analyses with colleagues. Foster collaboration and knowledge-sharing within your team. Annotate and comment on BLAST analyses (like in Acrobat). Share result with peers to leverage their expertise.
Export data in various formats
Streamline your R&D by integrating with other tools.
Download BLAST reports as HTML, XML or tab-delimited text. Programmatically submit and retrieve remote BLAST searches using python or command-line API. Import results into R or python. Export FASTA hit sequences or pairwise alignments. Link out to NCBI, Uniprot, jBrowse, IGV, or custom databases.
Custom links and design
Customize your SequenceServer and integrate it with other resources.
Add links from search hits to custom genome browsers or domain-specific databases. Integrate SequenceServer with other parts of your lab or community website. Change fonts, colors, and logos.
Don't melt your laptop!
SequenceServer Cloud supports intense and long-running queries.
SequenceServer Cloud is high-performance, high-availability, and scalable BLAST platform. It won't overheat (unlike your laptop!). Your BLAST search starts instantly. We intelligently leverage cloud computing to support large queries. Keep your laptop for lap-top-work.
Support your students
Intuitive BLAST analysis for large student cohorts.
No more waiting at the NCBI website. We leverage cloud computing to instantly start your BLAST search. SequenceServer intelligently scales to access (almost) infinite cloud resources. Hundreds of students can run BLAST seraches simultaneously. Perfect for teaching labs with large class sizes. Our intuitive user interface and visualizations flatten learning curves. Monitor student engagement with detailed logging.
Robust, Secure and private
Secured to the higest standards.
We prioritize your data's security and privacy. With advanced SSL/TLS encryption, proactive firewalls, intrusion detection, and exclusive Virtual Private Clouds (VPCs), we safeguard your data to the highest industry standards. Control access with popular Single-Sign-On (SSO) providers and track interactions with detailed audit trail logs. Trusted by academics, biotech, pharma, and agro-industry users worldwide, we're committed to protecting your data as if it were our own.
Use Cases
Comparative Genomics
Compare genomes across multiple species, identify conserved regions, protein domains, homologous genes, copy number differences, sequence differences, species, and evolutionary relationships. Deepen our understanding of the genetic basis of various biological traits and functions, and evolutionary histories.
Gene Curation and Annotation
Validate and refine gene predictions using SequenceServer BLAST searches against known gene sequences and protein domains. Identify potential inaccuracies in gene annotations and improve the qualities of genesets and genome assemblies.
Visual Analyses
Make graphs for visualizing and publishing. Get a range of BLAST plots, such as summary statistics, pairwise alignments and hit length distributions. Explore SRA read alignments, SNPs and INDELs with the IGV genome browser. Make and scrutinize gene and protein domain annotations with DNA Visualizer.
Design Diagnostic Markers
SequenceServer helps identify unique sequence regions in viruses, bacteria or other target organisms that can serve as diagnostic markers for disease detection. Design PCR or qPCR primers and probes, or specific guide RNA oligos (gRNA) for CRISPR/Cas9 assays. Improving the specificity and sensitivity of molecular-genomic approaches supports disease surveillance and management efforts.
Functional Characterization
Identify and analyze novel genes of interest in non-model organisms, as well as newly sequenced or understudied genomes. By comparing these sequences to known genes in other organisms, researchers can gain insights into the potential functional domains and biological roles of the newly discovered genes, guiding further experimental validation and functional studies.
Variant Calling Analysis
Explore DNA/RNA sequencing data from the NCBI Short Read Archive. Build SNP marker arrays for agricultural breeding. Identify and distinguish genetic variants in sequence data from wild species. Perform genome-wide mapping. Output alignment results for downstream analyses.
Data Hosting and Genome Browsers
Do you want to share genomes and annotations for your favourite species publicly or privately? SequenceServer makes sharing with others in your team or the wider community easy. Explore your data together with comments and genome browsers. Make your data have more impact.

Always Open Source
SequenceServer Cloud has many features that enhance productivity. But we started open source, and are proud to keep our core open source and free for those who want to run their own servers. Explore our documentation, connect with us our community support forum, or submit a PR to the GitHub repository. Together, we're advancing bioinformatics one BLAST at a time.
Ready to Revolutionize Your Sequence Analysis?
Get started with SequenceServer today
Try our Cloud ServiceLatest Updates
2025-12 BLASTing to get 1000s of hits