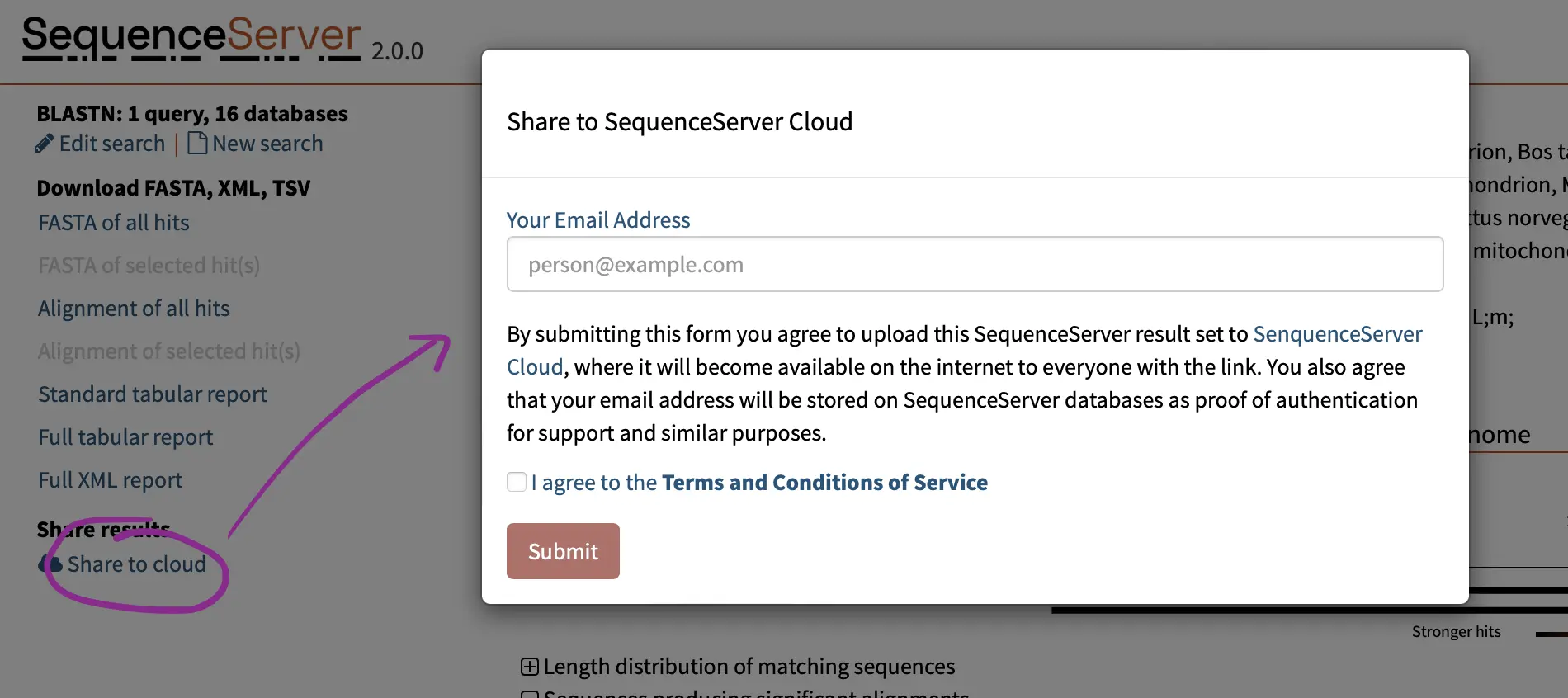
Overview of SequenceServer Features
SequenceServer has many features that will accelarate your research,
allowing you to extract the most from your data.
Powerful BLAST Submission Interface
SequenceServer provides an easy-to-use and helpful interface. It auto-detects whether the input sequence is either nucleotide or amino acid. With the combination of input data and database type, SequenceServer selects only appropriate BLAST algorithms to use. This helps you to make mistake-free analysis of your data.
SequenceServer comes packaged with useful databases as default. These include NCBI nr/nt and UniProtKB/SwissProt databases, and genomic, transcriptomic and proteomic databases for several of the most commonly searched organisms (human, mouse, fruit fly and thale cress). This saves time, effort and database management.
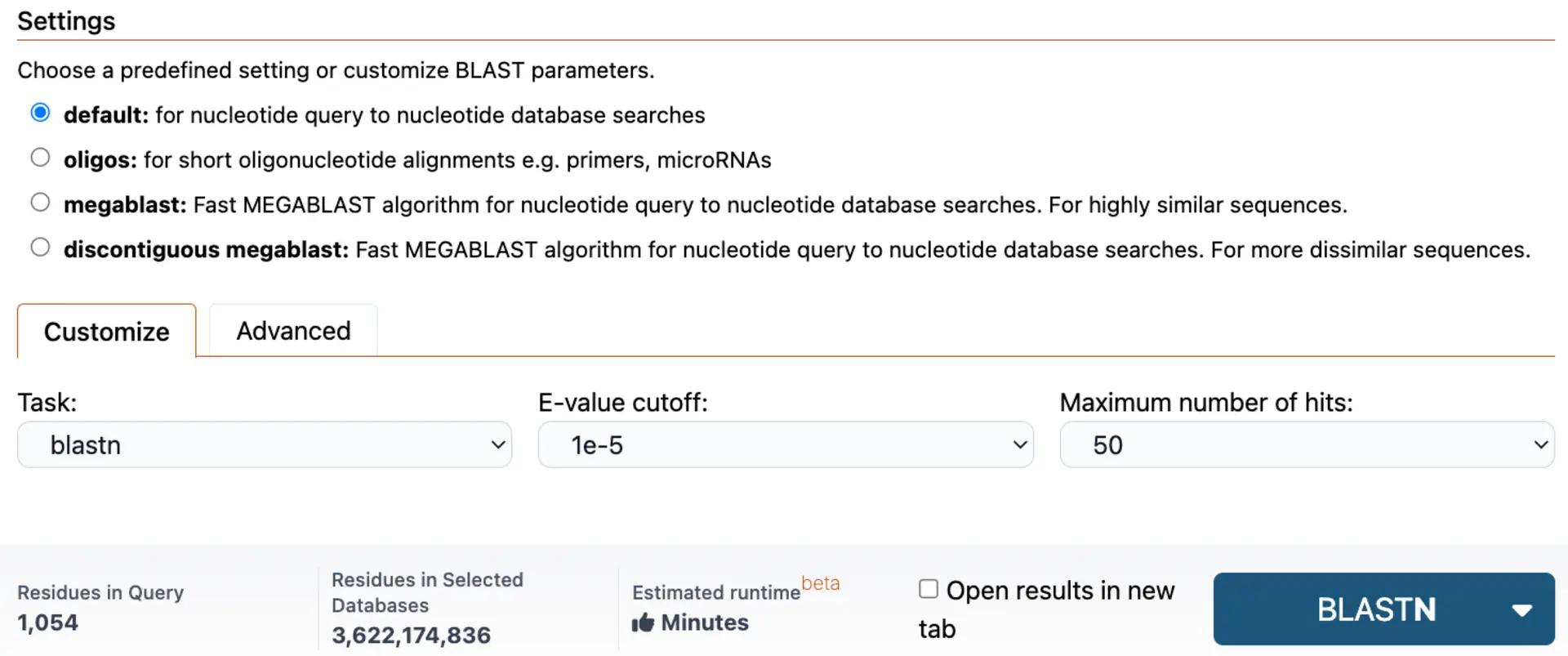
East-to-Use Advanced Parameter Selection
SequenceServer gives you access to preset parameter options, enabling you to analyze different data types. Alternatively, you can have complete customization of parameters, with your parameter history saved for future use. This flexibility enables you to optimize your time and search sensitive.
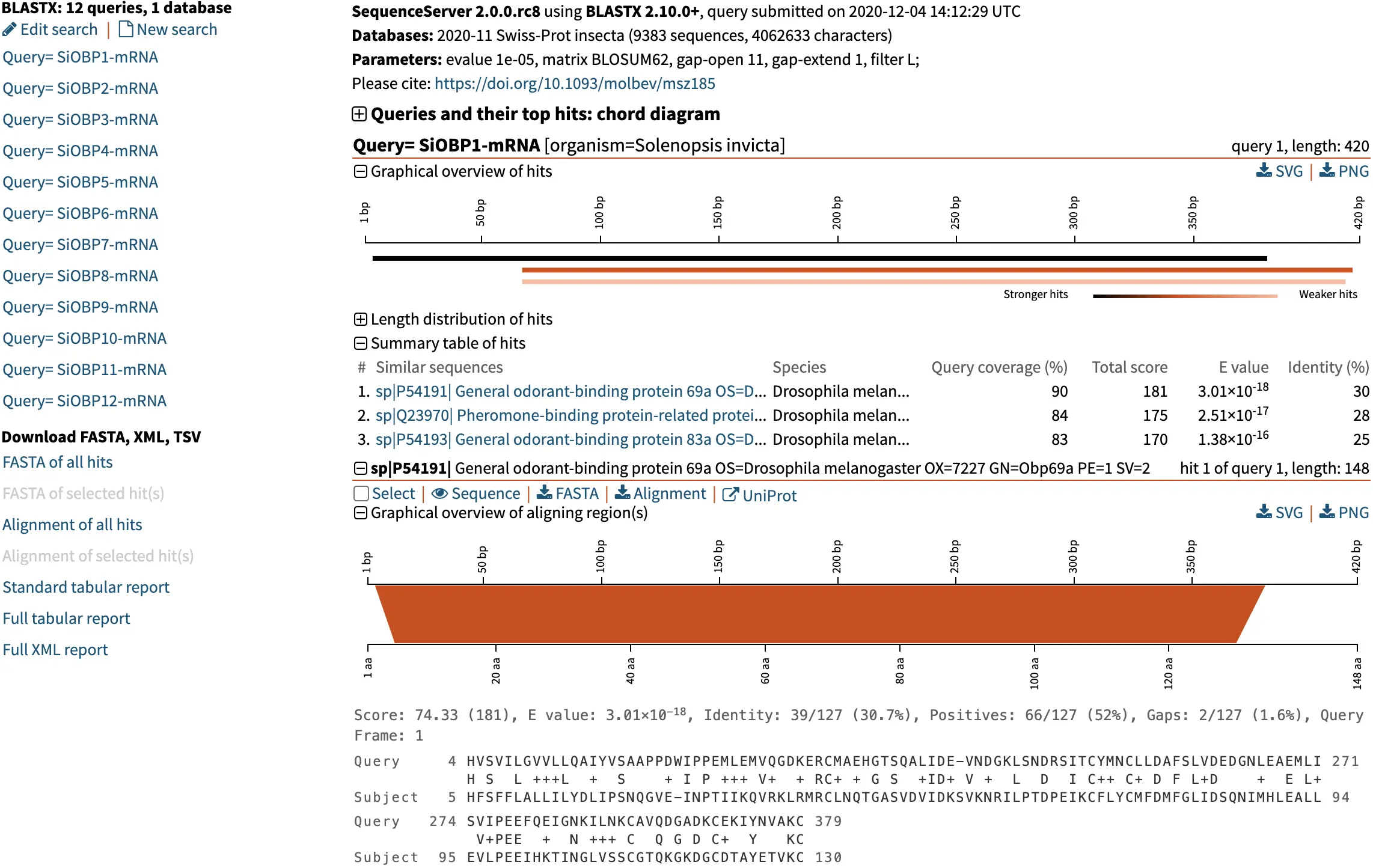
Expansive BLAST Result Overview
SequenceServer goes beyond your standard BLAST results. A comprehensive overview of your search results is generated in an interactive and easy-to-manipulate format. Multiple different graphical representations of results allow you to visualize and interpret BLAST results. These graphs can be easily downloaded and are ideal for publication and presentation.
SequenceServer provides a range of functionality tools, allowing you to extract reports, alignments and FASTA files.
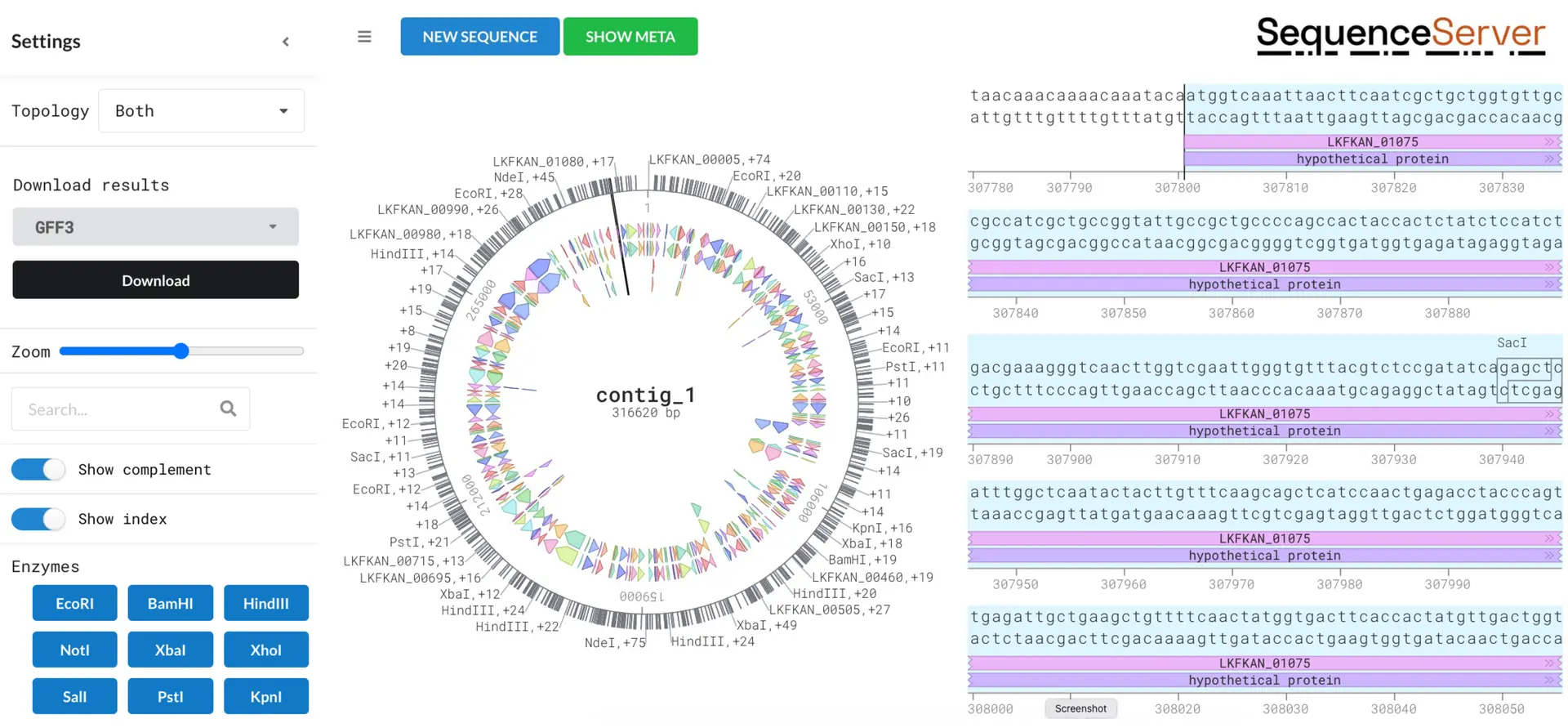
Annotate and explore features with DNA Visualizer
SequenceServer provides a straightforward bacterial genome and plasmid annotation tool. This makes gene identification and exploration quick and easy. The interactive interface allows for customization of plots so you can focus in on the data that matters. Outputs for downstream analysis are easy to download.
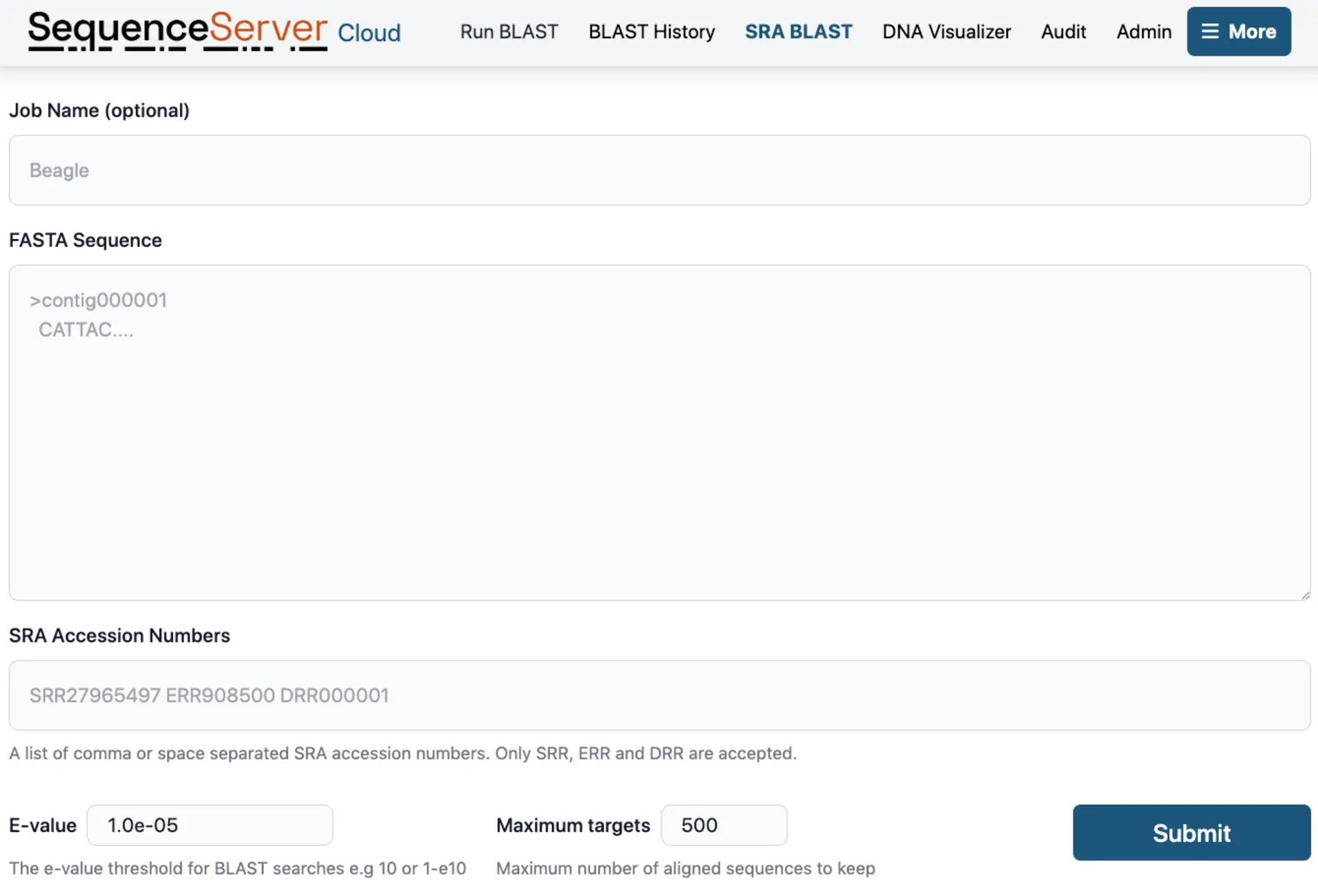
BLAST to the NCBI Short Read Archive
SequenceServer enables you to search the NCBI Short Read Archive (SRA) with ease. This is a powerful way to explore a huge repository of data without the need for download times. SequenceServer allows you to download various formats including alignments, FASTA files of hits, and BLAST results output, which facilitates further downstream analysis.
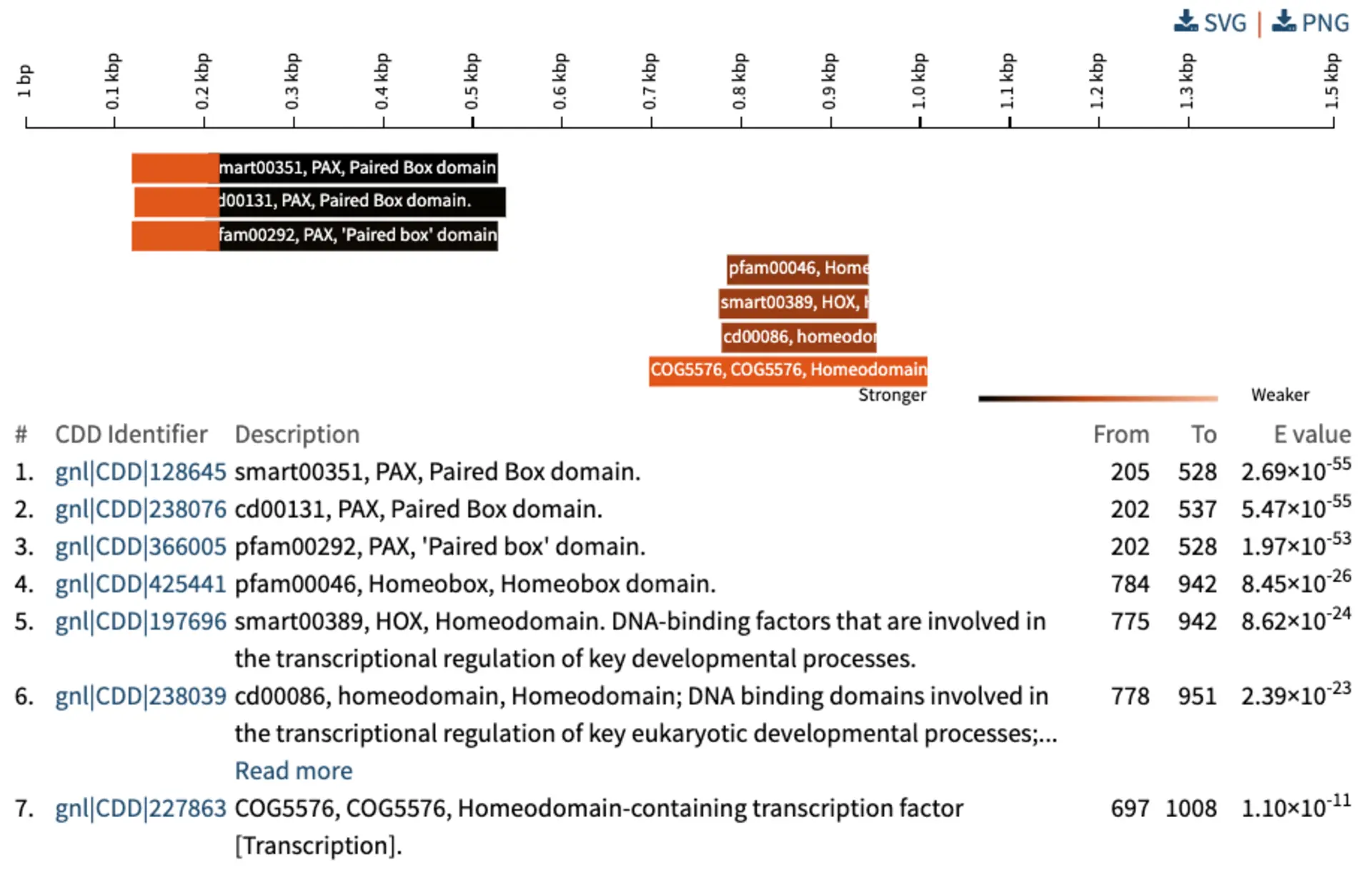
Analysis of Conserved Domains
SequenceServer identifies possible functionality by annotating your query and BLAST hits using the Conserved Domain Database (CDD). This provides immediate help in the identification of possible orthologues and functional similarities during your BLAST searches. Graphical and text outputs are provided so that you can understand and analyze your results how you prefer.
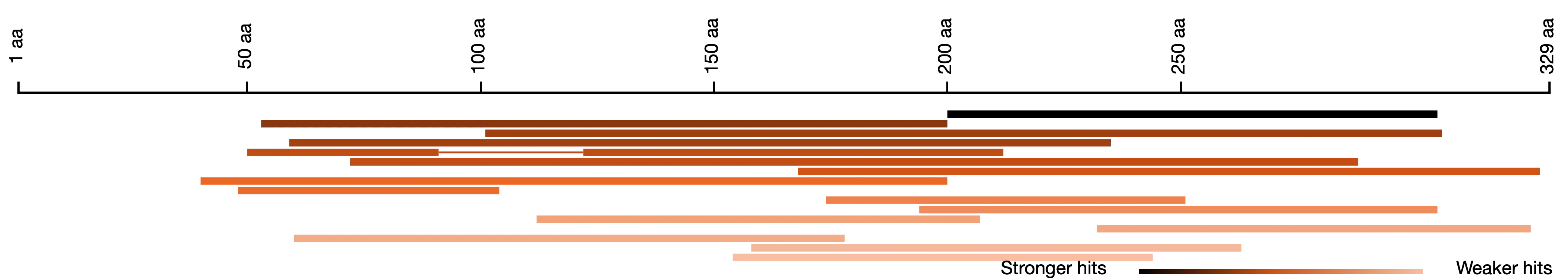
Length Distribution of BLAST Hits
Length distribution and identity metrics of your hits help to identify similarities and differences between your query and the BLAST hits. For example, this query (a predicted gene product) is approximately twice as long as the sequences it is similar to.
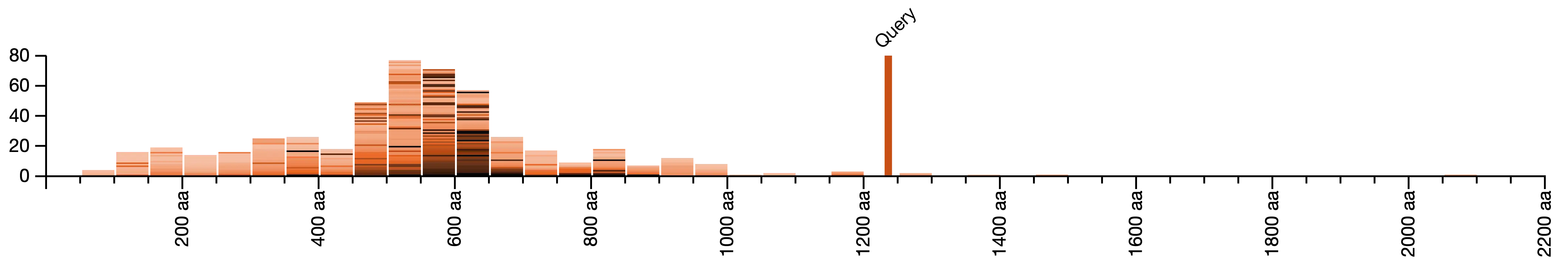
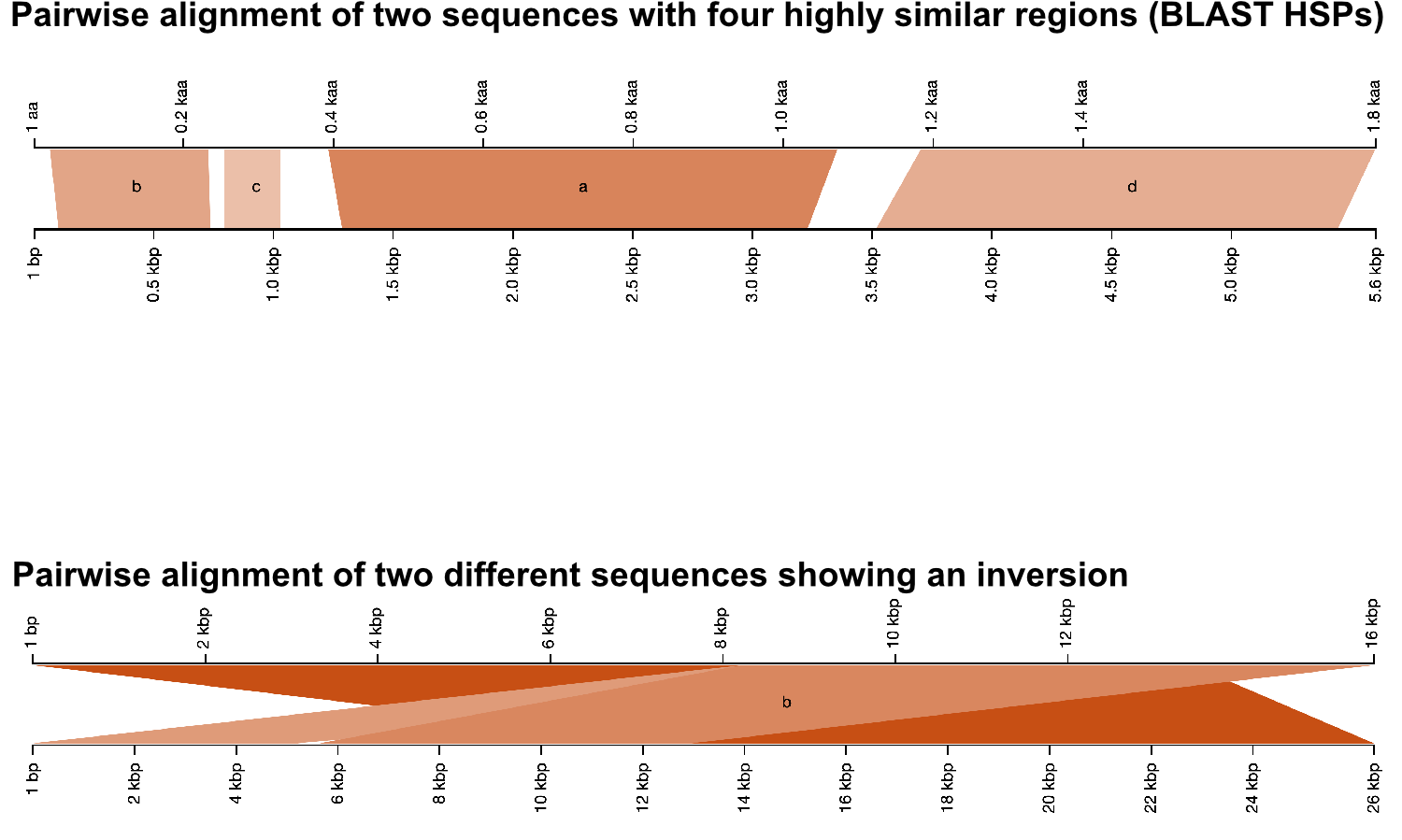
Pairwise BLAST Alignments
Visualization of pairwise BLAST alignments is a powerful means to quickly understand how your query aligns to hits. In combination with SequenceServer’s CDD annotation, this enables you to see what regions of your alignment might relate to important conservation and divergence.
It gives quick interpretations of your alignment, allowing you to detect and explore genomic features like inversions, insertions and deletions.
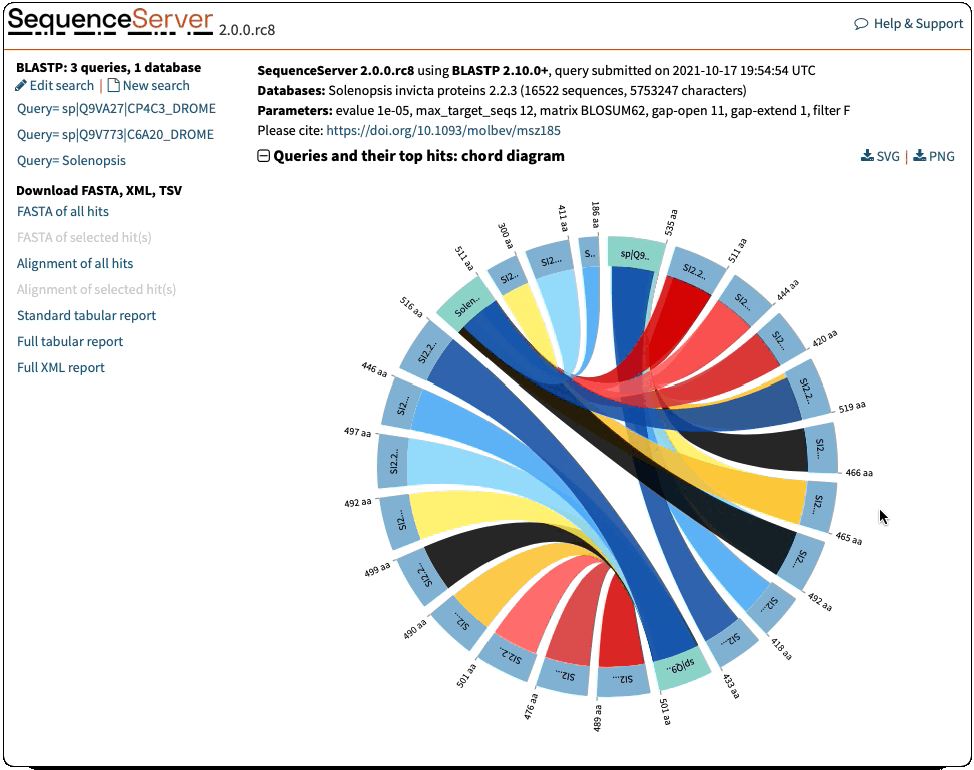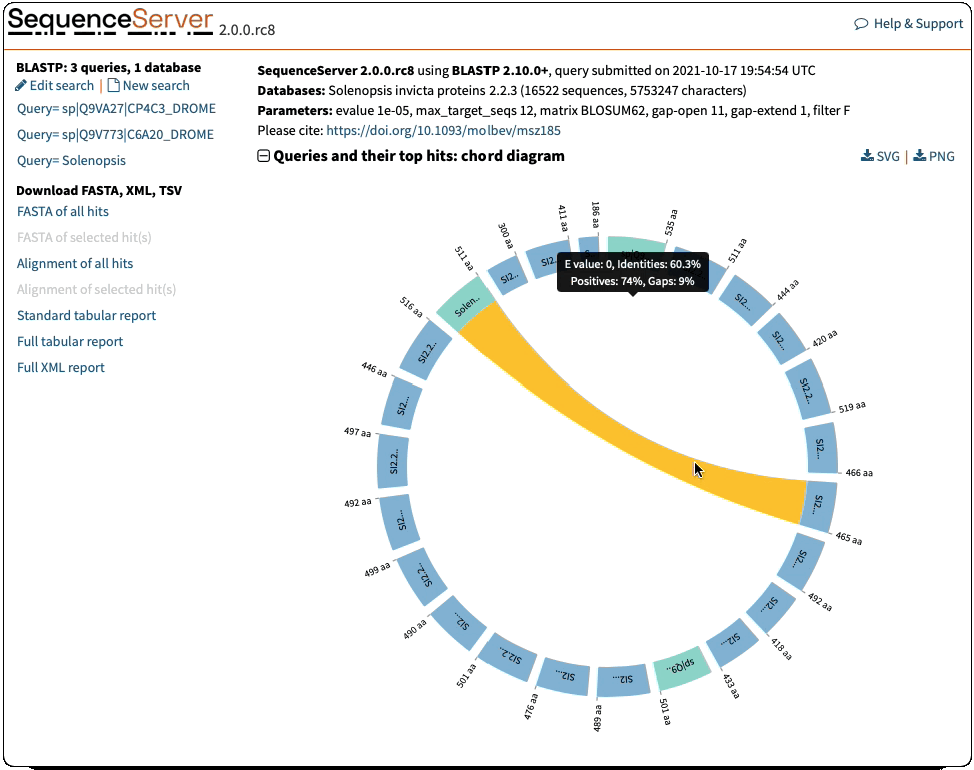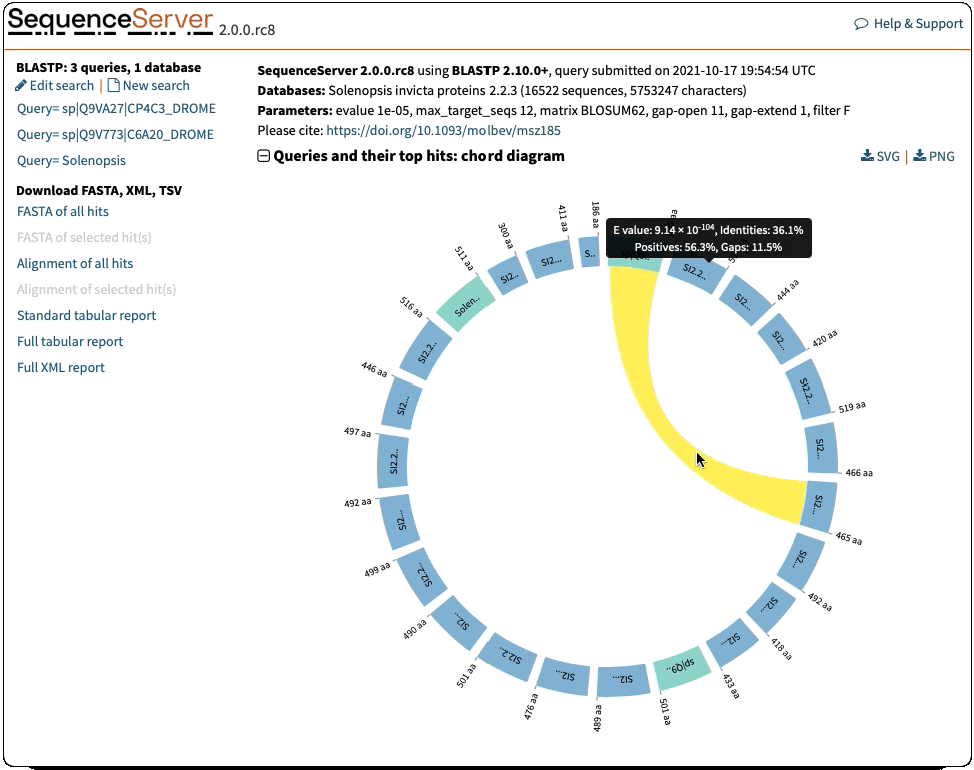
Circos-Style Visualization of BLAST Queries and Hits
Offering a compelling and intuitive representation of BLAST queries and hits. These plots are a great way of interpreting multiple hits simultaneously. They give information of the score (E-value), position and length of alignments. Interactive elements allow you to select the features you want to highlight.
BLAST Hit Similarity Score
Get an overview of where alignment hits fall in relation to your query. BLAST hit sequences (shades of red) aligning to the query sequence (black). Darker color means stronger similarity (i.e., E-value).
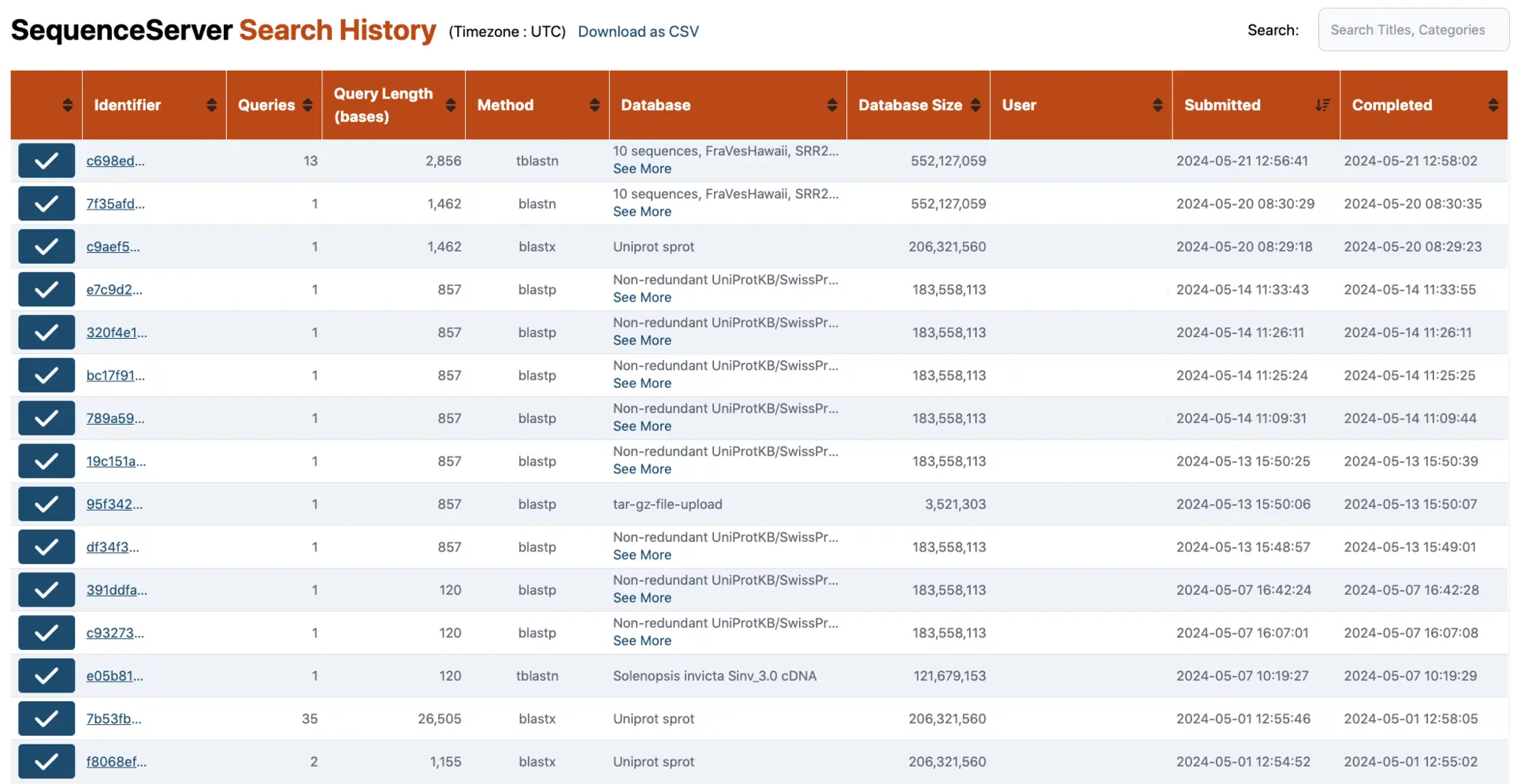
History of BLAST Searches
Never lose track of your research with SequenceServer’s automatic logging and retrieval. You will always be able to go back to your previous BLAST results. This ensures you can keep track of software parameters, datasets, queries and more, making your research robust and reproducible. This overview page of BLAST searches done by you and your team gives you access to all previous results.