BLAST & Genome Bioinformatics thoughts
We think constantly about improving genomic analysis.
BLASTing to get 1000s of hits
Max hit number
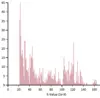
Histograms of general BLAST statistics
Summary statistics histograms for BLAST analyses provide an easy-to-interpret visualization method to extract meaning from BLAST searches.
Interpreting BLASTN nucleotide BLAST results
Exploration and overview of BLASTN inputs and outputs. This covers the core concepts and metrics used with BLASTN
Taxonomic restriction of BLAST searches
How to taxonomically restrict BLAST searches to achieve better results and refine your research questions
DNA feature exploration with the DNA Visualizer SequenceServer tool
Detection and visualization of DNA features. An easy-to-use bacterial genome and plasmid annotation tool, with added visualisation tools.
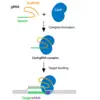
Generating guide RNAs and sequencing primers for CRISPR knockouts
How to use SequenceServer to design guide RNAs for CRISPR knockouts, assessing mutation sites and off-targetting
Search Raw Reads from NCBI's SRA Database 🧬
How to search the SRA Database with SRA BLAST. Enabling massive screening of publicly available data
Improvements to BLAST search in SequenceServer 3.0
Release notes for SequenceServer 3.0. This release includes new features, maintenance, bugfixes and minor improvements.
Similarities, differences and usage contexts of FASTA and FASTQ sequence formats
What are FASTA and FASTQ sequence formats, when are they used, and what kind of information do they contain.
Citation Guidelines for NCBI BLAST
It's important to cite BLAST and its related software and data sources in your publications.
Identifying conserved protein domains to understand gene function
Annotate proteins with the CDD Conserved Domain Database in SequenceServer. Improve your understanding of functional domains in your sequences
Referral program: help your friends and get paid!
We've started a referral scheme. Help your friends and earn cash!
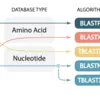
BLASTN, TBLASTX, BLASTP, TBLASTN, BLASTX - which should I choose?
BLAST algorithm choice depends on what type of query sequence you're comparing to which type of database. It's critical to choose the correct one.
Get your exclusive SequenceServer stickers – and more!
Free stickers for SequenceServer users! And enter a drawing for surprise swag!
Updates, fixes, and improvements for SequenceServer BLAST 2.2.0 - release notes
Version 2.2.0 starts up much faster, and improves performance and usability with large queries
Historical overview of BLAST searches and optional logging and tracking usage through detailed audit trails
Audit trails and access trails for BLAST database. Great for monitoring student engagement, and Configuring user access and databases for a SequenceServer Cloud BLAST server
Setting Up and Running a Custom BLAST Server: technical & logistical considerations
Appropriately leveraging powerful tools like BLAST is essential for success. Here, we walk through some of the things you need to think about when setting up and running a custom BLAST server.
Updates, fixes, and improvements in SequenceServer BLAST 2.1 - release notes
Version 2.1 includes many new features and bugfixes, including Cloud Sharing, NCBI BLAST 2.14, backend improvements and improvements to architecture and robustness.
Disentangling homology, orthology, paralogy and similarity with BLAST
We define what are orthologs, homologs and paralogs. Using examples and a case study (Tachykinin vs Neuro-peptide Y) we clarify how BLAST can help to elucidate relationships among genes.
Metabarcoding to identify organisms from environmental DNA (eDNA) with BLAST
BLAST can help identifying organism in an environmental sample using a metabarcoding approach
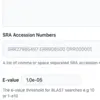
How BLAST E-values are calculated and what they mean
Here we take a look at the BLAST E-value and how it is used in BLAST
Identifying gene sequences in a new genome assembly using BLAST
BLAST can help us obtain the gene sequences from the genomes we are interested in
Check PCR primer specificity using BLAST but without primer blast
Five things to do when checking primer specificity using BLAST. Make sure you get the best PCR results!
Using FASTQ format sequence reads to run BLAST searches
BLAST is powerful, but not necessarily appropriate for analysis of FASTQ files
BLAST in the cloud
Run BLAST analyses in the cloud to get results faster, or search on unpublished data.
Understanding and interpreting BLAST alignments is easier with visualizations
SequenceServer incorporates four powerful visualizations that help researchers interpret BLAST alignment results.
Point and click BLAST server configuration
How to configure user access and databases for a SequenceServer Cloud BLAST server.
Exploring the methods of sequence searching with SequenceServer BLAST
Whether you are studying the genetic makeup of an organism or trying to identify potential targets for drug development, sequence searches can provide valuable insights and information.