Configuring user access and databases for a SequenceServer Cloud BLAST server
Many SequenceServer users simply see the graphical interface for performing BLAST analyses.
But how do you format FASTA files from genomes or transcriptomes into BLAST databases? How do you make your server secure so that only certain people can access it? If you’re running your own server, most of this happens in the command-line.
However, if you’re using SequenceServer Cloud just use the graphical interface to configure your server. The command-line is not needed.
Administration dashboards for a SequenceServer Cloud BLAST server
Each SequenceServer Cloud BLAST instance has a point-and-click administration dashboard. This dashboard enables you to:
- Add, remove or rename databases
- Add or remove users
- Change your subscription and billing information
The administration dashboard of each SequenceServer Cloud instance is accessible at https://myinstance.sequenceserver.com/edit. It has three sections:

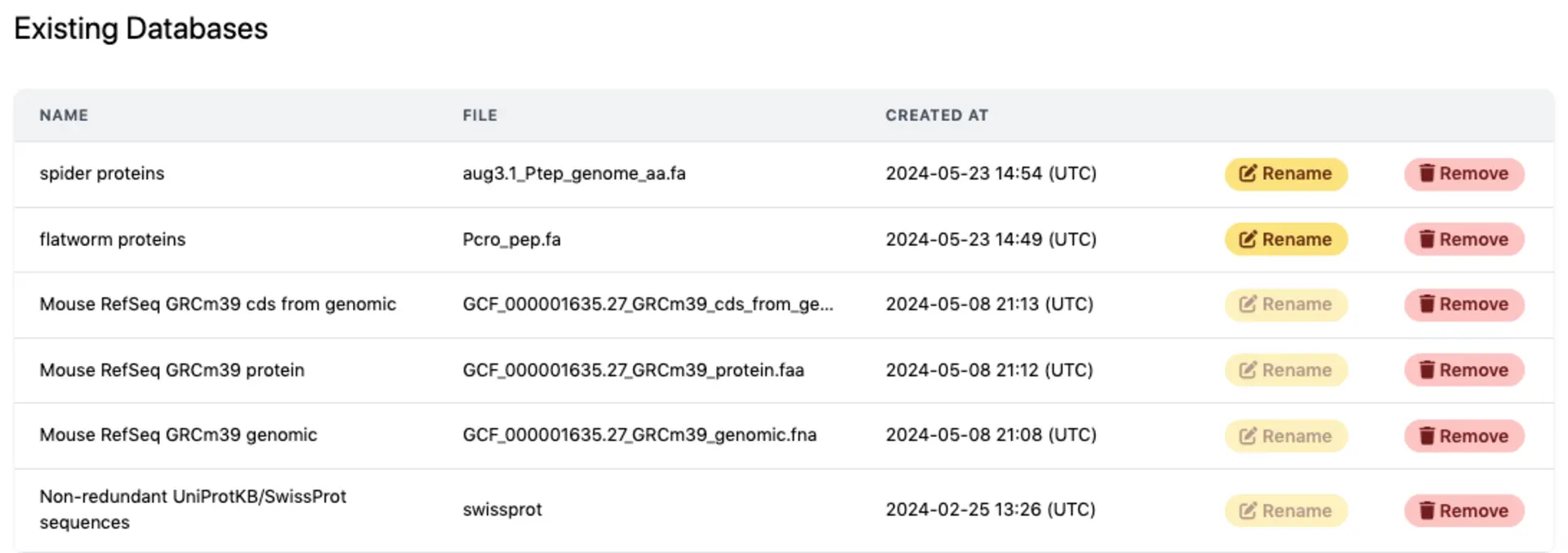
Overview of existing databases
The database section includes an overview of existing databases, and options for adding more.
Options for adding BLAST databases
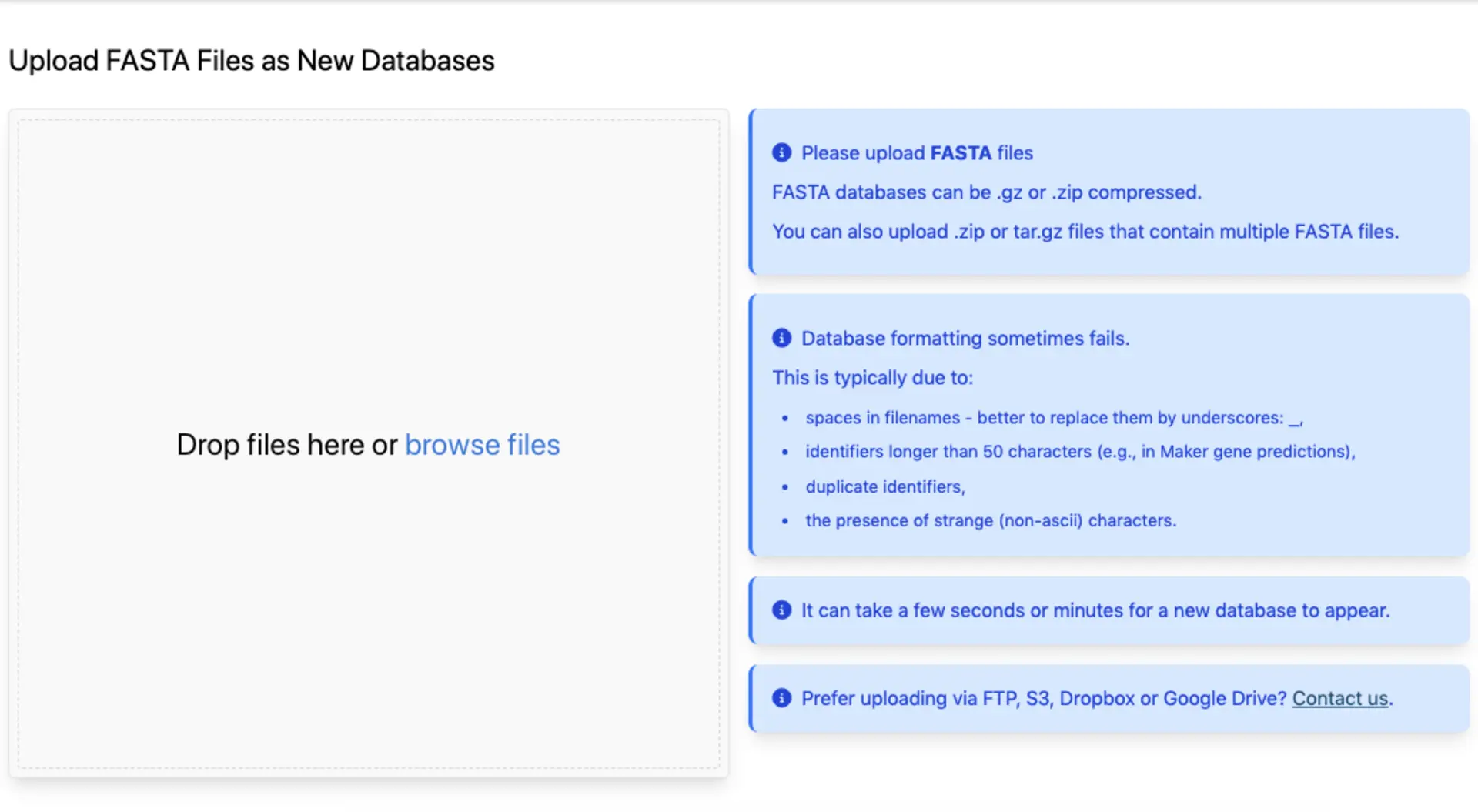
Adding custom databases
To add your own data you can drag and drop (or select) individual FASTA files or gzipped FASTA files, or provide URLs to such files. You can also batch upload many FASTA files as a single .zip or .tar.gz. We automatically detect whether they are protein or nucleotide FASTA files and format them into BLAST databases.
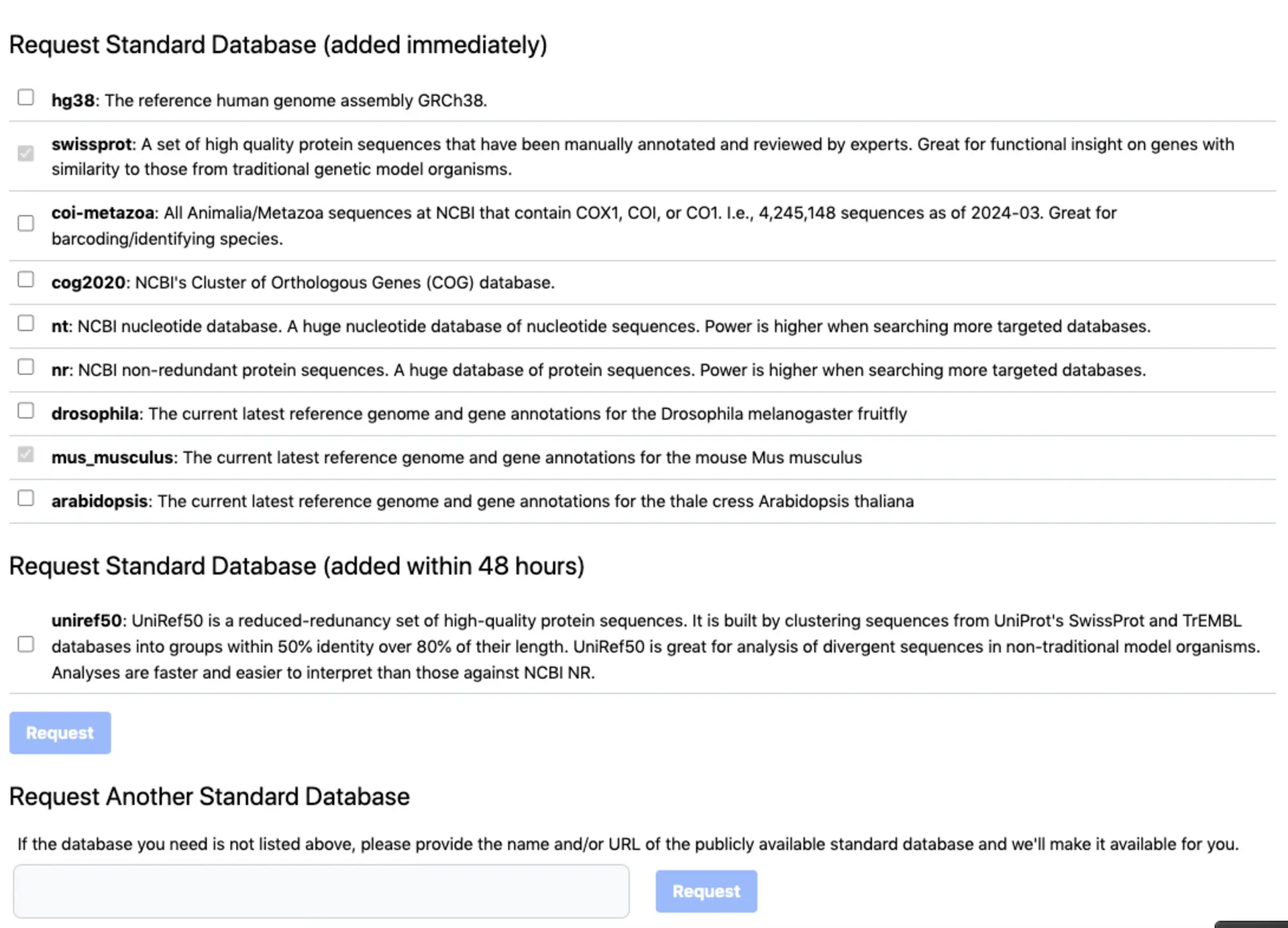
Adding commonly used databases
You have access to commonly used databases that come with SequenceServer. These include datasets for model species, and NCBI and SwissProt. You can also request other databases to be added.
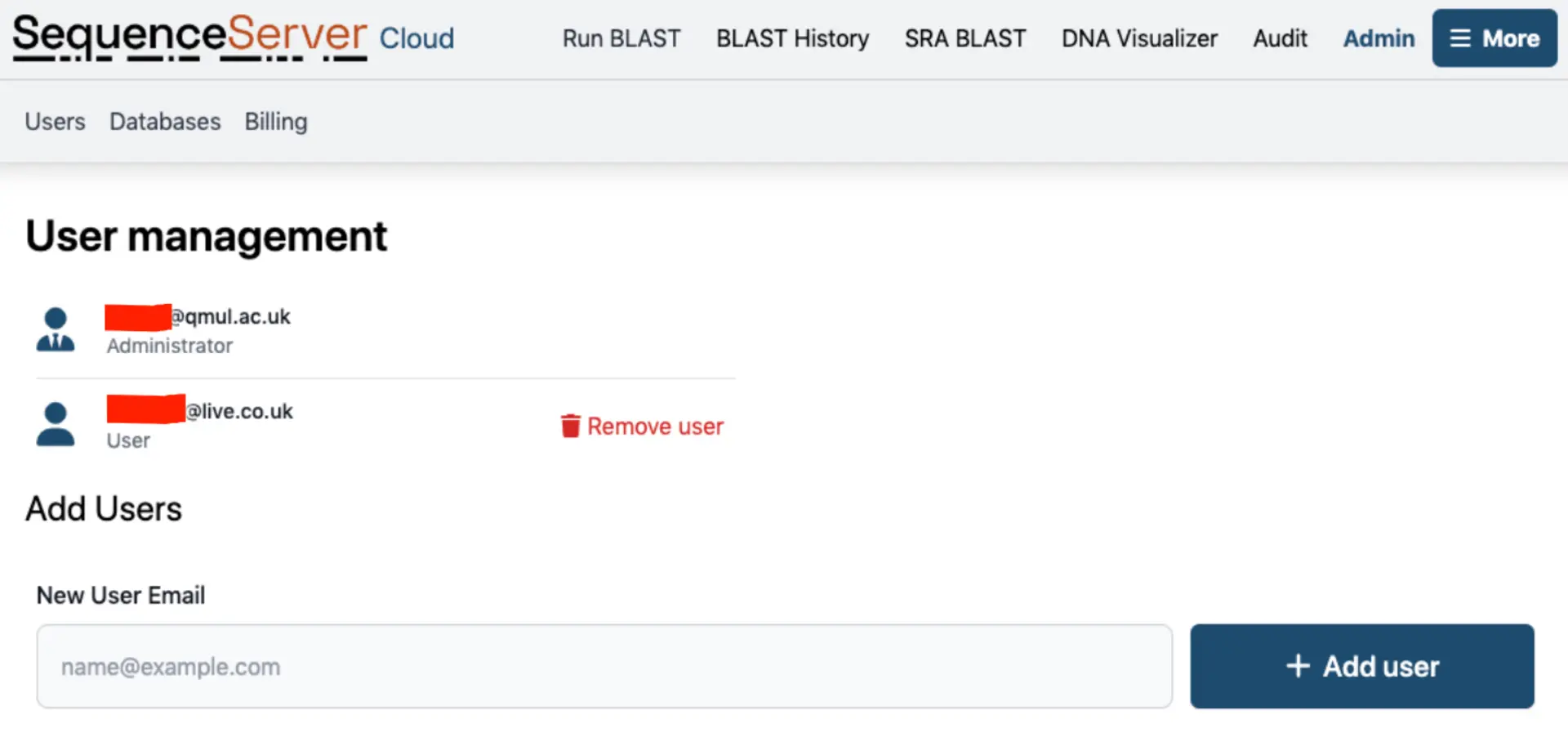
Adding Users
If your SequenceServer Cloud instance is private, you can grant access to colleagues in the “Users” tab by entering their emails and clicking “Add user”. They will receive an email with a link to the BLAST interface.
When a user tries to access the BLAST interface, they will be asked to enter their email address (left), and will receive an email with a magic login link (right).
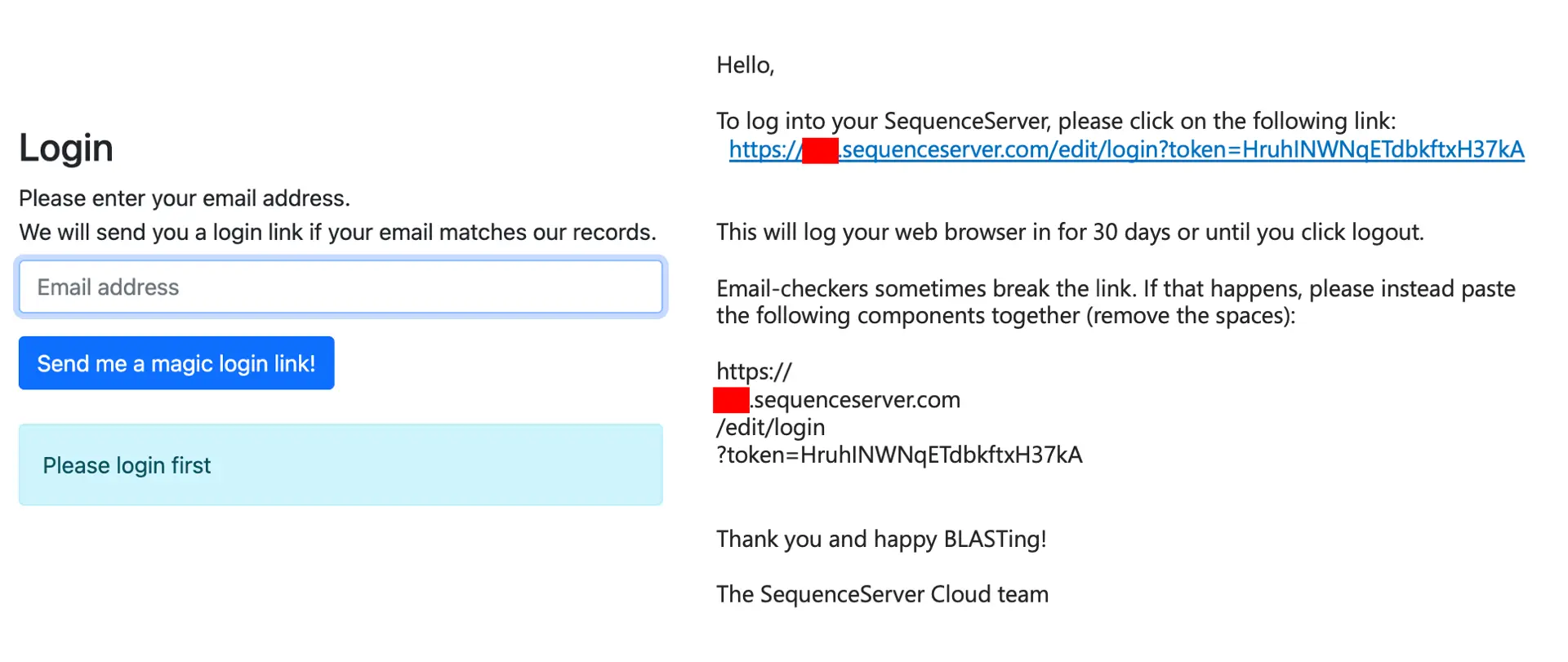
SequenceServer instances can also be publicly accessible. In that case, no user-specific things are required.
Some organizations prefer using their Signle-Sign-On (SSO) login, as provided by AzureAD, Okta, Google or other. We can work with most SAML identity providers. Just get in touch. Other organizations like additional security though, for example restricting access to particular IP addresses, or using a generic http password. Contact support if you’re in either situation.
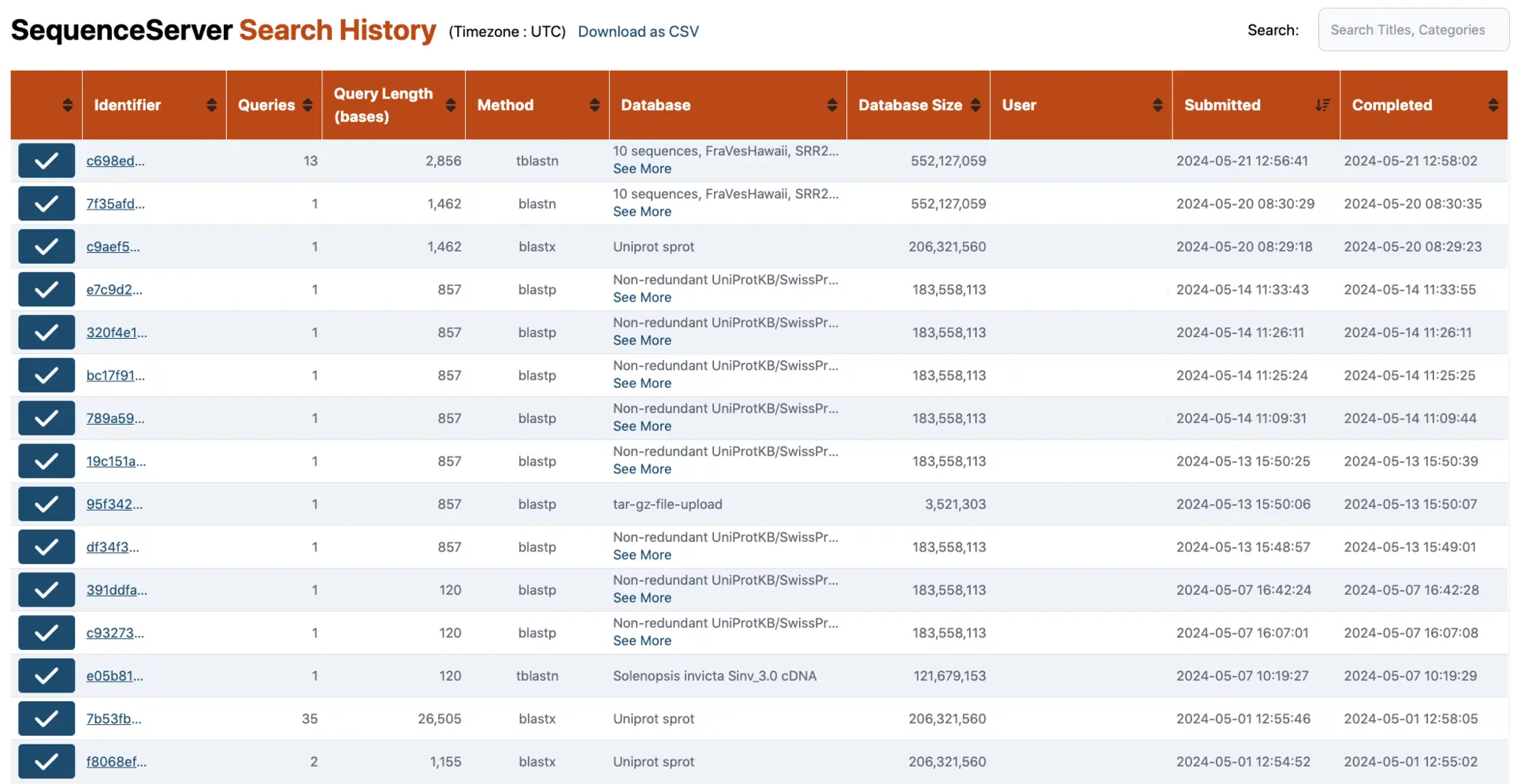
BLAST search history
By default, a button at the top of the dashboard allows the administrator(s) to view the search history of all users. We can also make this accessible to all users.
This overview of searches performed can help researchers to go back to a previous result, and can help administrators to understand which datasets are most important to users.
Advanced logging
Advanced logging is also available. We can activate “audit trail logging” whereby every click is recorded. This can help with security audits, and for better understanding which of your team members are using the server and how. This is also great for teaching, as it can help educators make sense of student engagement.
By leveraging cloud computing, publication-ready graphics, and a powerful administration interface, SequenceServer Cloud makes it easy to perform sequence search results and to interpret them.
