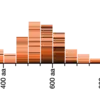SequenceServer BLAST Release Notes
SequenceServer v3.1.3
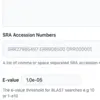
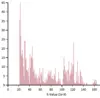
We’ve added histograms of BLAST analysis statistics, and made several improvements to SRA BLAST.
There are also many less visible improvements & fixes:
- Refined JavaScript Handling:
- Split sourcemaps from minified JS for cleaner builds.
- Refactored inline JS to better comply with Content Security Policies (CSP).
- UI & UX Enhancements:
- Fixed footer styles and addressed flickering issues in results display.
- Switched input fields to a monospaced font for improved readability.
- Security, Validation & Stability:
- Updated JavaScript dependencies and bumped
nokogirifor added robustness. - Implemented more thorough input validations to ensure data integrity and reduce errors.
- Updated JavaScript dependencies and bumped
- Functional Adjustments:
- Adopted
blast 2.16.0. - Changed the download label from “Pairwise” to “Text”.
- Enabled advanced parameters to always be rendered.
- Rendered JSON errors correctly on
.jsonrequest paths. - Considered path configuration when extracting tasks.
- Adopted
v3.1.1
Notable Updates:
- Downloads & Content:
- Enabled downloading results as pairwise TXT.
- Added more stringent download validations.
- UI & Security:
- Fixed the 404 page logo display issue.
- Introduced CSRF protection for enhanced security.
- Infrastructure:
- Transitioned to using Rackup.
- Replaced
batchwithentry_batchfor improved clarity.
2024-06-12 SequenceServer BLAST 3.1.0
This release contains both user-facing and technical changes that you can explore in detail on GitHub.
New Features and Enhancements
- Fresh new UI for the query page: still familiar, but with significant improvements and more modern look. Admins can now add human readable descriptions for pre-defined BLAST search parameters that are displayed as radio buttons. The call-to-action button sticks to the bottom of the screen if the SequenceServer instance contains a long list of databases or predefined options that used to push this button below the fold.
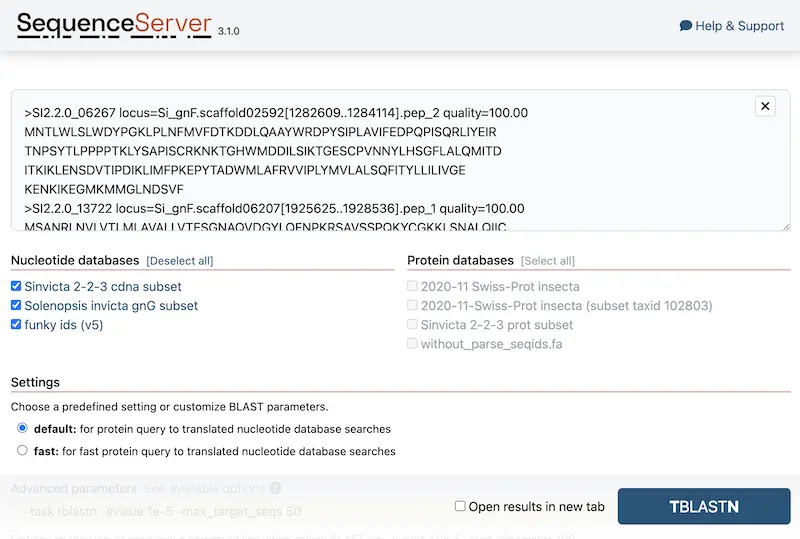
-
Remember section collapse state: we’ve realized that sometimes users only care about specific sections of result pages. To make it more delightful, SequenceServer now remembers what sections you’ve collapsed before and retains the same visibility settings across all result pages.
-
Release notifications: checks if a new version of SequenceServer is available and displays an unobtrusive notification in the UI about it.
-
Improved query type detection: SequenceServer is now smart enough to skip all unknown bases when detecting the user query type.
Maintenance Updates
These are some technical changes that help make things faster, more reliable and easier for us to change things in the future.
-
Dockerfile now uses BLAST 2.15: thanks to a contribution from Nathan Weeks.
-
SequenceServer requests BLAST 2.15: SequenceServer has been compatible with 2.15.0 since 3.0 without any issues, bump BLAST dependency to 2.15.0+.
-
Improved CI pipeline: less noisy and easier to look after configuration.
-
Backend code refactoring: Code simplification to make SequenceServer easier to extend in the future.
-
Reduced reliance on jQuery and Bootstrap CSS: The new query UI uses Tailwind CSS that allows iterating our UI faster. We’re also removing reliance on jQuery in favour of modern JS and React.js for similar reasons.
-
Number of sequences in a query: record the number of FASTA sequences in a query in job.yaml.
Like always, SequenceServer Cloud users will have these and many more changes available on their instances starting today.
2024-02-14 SequenceServer BLAST 3.0
This marks a significant update to SequenceServer, introducing a plethora of features, improvements, and bug fixes aimed at enhancing the bioinformatics analysis experience. The full list of commits and detailed changes can be accessed on the GitHub repository.
New Features and Enhancements
-
BLAST Database Formatting Errors: Improved error messages for BLAST database formatting issues, such as duplicate identifiers or illegal characters, making it easier for users to diagnose and fix database issues.
-
FASTQ Support: SequenceServer can now automatically detect FASTQ files and convert them to FASTA format, offering greater flexibility in handling different query input types.
-
Extended Query Size: We have expanded the maximum size of input queries from 10Mb to 250Mb, allowing for more extensive BLAST searches suitable for larger datasets.
-
Cloud Service Notifications: SequenceServer Cloud users will now receive email notifications upon the completion of long-running BLAST jobs, adding convenience and improving workflow.
Maintenance Updates
-
Dependency Management: Major updates have been implemented in our software dependencies to ensure continued reliability and security. The Sinatra framework has been updated from version 2 to version 4, and the corresponding Rack dependency has moved from version 2 to version 3.
-
Encoding Standards: UTF-8 is now the default encoding to ensure consistency and compatibility across different computing environments.
User Interface and Usability Improvements
-
Results Page Enhancements: Adjustments and optimizations have been made to the results page, including CSS paragraph clamping and making gzipping of responses optional.
-
Initialization Robustness: A more robust initialization process that gracefully handles cases with unformatted databases.
-
Dedicated 404 Page: A dedicated 404 page has been added to handle missing pages and broken links more elegantly.
-
Sidebar Navigation: Improved sidebar element clickability for a more intuitive user experience.
Performance Optimizations
- BLAST+ 2.15: Integration of the latest BLAST+ version brings performance improvements and new functionalities to SequenceServer.
Security and Stability Fixes
-
404 Exception Handling: Fixed an issue where job 404 exceptions were not handled properly, enhancing system stability.
-
Browser Compatibility: Dropped reliance on the webshim polyfill, ensuring better performance and compatibility with modern browsers.
Small List of Changes
- Results Page Tweaks: Several adjustments for improved display and interaction with BLAST results.
- Encoding and Performance: Transition to UTF-8 as the default encoding and optional gzipping of responses to optimize loading times.
- Cloud Notifications: Email notifications system implemented for SequenceServer Cloud services.
- Other minor changes… see the Full Changelog.
2023-10-04 SequenceServer BLAST 2.2
Noteworthy Feature Enhancements
-
Speedier Launch Times: Thanks to two significant changes, SequenceServer now boots up much faster. By deferring database formatting checks to only when necessary and introducing the
--optimisticflag, which bypasses all checks, users can experience rapid startup times, especially on systems with extensive databases. -
Handling of Large BLAST Results: For analyses resulting in extensive data (e.g., BLAST outputs exceeding 250Mb), SequenceServer offers a solution to avoid browser and server overload. Instead of attempting to render massive result pages, a warning is issued, and users are given the option to download the results in .xml or tab-delimited formats.
User Experience Improvements
-
Format Recognition for .faa Files: To ensure compatibility and user convenience, SequenceServer now automatically recognizes and formats .faa files, a less common extension used for amino acid sequences in FASTA format.
-
Enhanced Error Tracking: Database formatting errors are now tracked and logged, providing clearer feedback for troubleshooting and enhancing overall user experience.
Infrastructure and Compatibility
- Docker Improvements: Upgrading the Docker build environment to
ruby:3.2.2-bullseyefrom the previousslim-busterbase image has improved overall compatibility and set the stage for future enhancements. - Architectural Rewiring: Codebase tweaks have been implemented, resulting in improved maintainability and setting the stage for future development and features.
- Cloud-Specific Advancements: Several features and improvements have been made exclusively available to users of SequenceServer Cloud BLAST, demonstrating our commitment to providing a premium experience for our cloud service subscribers.
2023-08-01 SequenceServer BLAST 2.1
User Experience and Collaboration
- Upgrade to BLAST 2.14: Access to faster algorithms and improved analysis speed.
- Cloud Sharing: Introduced cloud sharing of results pages via a secret URL for easier collaboration.
- Documentation Improvements: Enhanced documentation for optional BLAST parameters.
- Enhanced Performance: Improved handling of slow connections and server response times.
Code Quality and Security
- JavaScript and Ruby Upgrades: Transitioned JavaScript code to ES6 and upgraded to Ruby 3.2 for improved security.
- Testing and Security Enhancements: Introduced JavaScript unit testing and strengthened security measures.
- Continuous Integration: Shifted from TravisCI to GitHub actions for automated testing.
2020-12-11 Many improvements due to Hacktoberfest and other initiatives
We are pleased to announce the next candidate release of SequenceServer 2.0.
- Automatically check for incompatible databases on startup and prompt users to reformat them.
- Upgrading from the older (V4) to the newer (V5) database now preserves any taxonomy information embedded in the database.
- Add ability to use -taxids_map of makeblastdb during database creation. To use it, add a ‘.taxids_map.txt’ file next to the FASTA file (e.g. “/db/ants.taxids_map.txt” for “/db/ants.fa”)
- Add option in the search form to open BLAST results in a new tab.
- Add ‘Edit search’ and ‘New search’ links to the report page. (thanks to Tomas-Pluskal & TomMD for the push)
- Search form can now be cleared by reloading the page. Relevant if you use your web browser’s back button or ‘Edit search’ link and want to clear the form to start over.
- Make it easier to pass command line arguments to Docker image. For example, number of BLAST threads can now be set as “docker run … wurmlab/sequenceserver sequenceserver -n 4” instead of “docker run … wurmlab/sequenceserver bundle exec bin/sequenceserver -d /db -n4” (yeah!)
- Reduce size of Docker image 3-fold. (credit: Nathan Weeks, Neel Kamath)
- Reduce size of gem file 3-fold. (credit: Nathan Weeks)
- Fix a bug that prevented query textarea from being resized veritcally. (credit: Iain-S)
- Fix a bug that prevented search form from rendering query sequences sent to it via POST request. (credit: Sam Hokin)
- Fix a bug that prevented the use of keyboard shortcut ‘Ctrl-Enter’ to submit the search form.
- Fix a bug that would cause an infinite loop on search form if the user had only one database.
- Fix a bug that could cause new users to be prompted more than once to join the announcements list.
- Fix a bug that was causing Travis builds to fail.
Special thanks to Iain-S for introducing us to hacktoberfest. It was incredibly productive, leading to changes 8-10 above.
2020-06-23 Support for NCBI BLAST’s v5 database format
Based on your feedback we have fixed bugs affecting docker and sub-URI deployments, updated documentation for HPC-integration, and most importantly, we have created a path for migrating to NCBI’s newer V5 database format (see release notes below).
- Running ‘sequenceserver -m’ will now automatically detect older V4 databases, and those created without the ‘-parse_seqids’ option of makeblastdb, and offer to rebuild them. This works even if you deleted the original FASTA files. Database titles are preserved when rebuilding, however, taxonomy information in the database is unfortunately not preserved.
- If you do not wish to rebuild your databases yet, you can now run SequenceServer with BLAST+ 2.9.0 instead. The tradeoff is that you would have to manually download BLAST+ 2.9.0 and indicate its location in the configuration file. SequenceServer will only download 2.10 by default.
- If building BLAST databases fails for some reason, the command that was tried and the resulting output is properly relayed to the user so that the problem can be diagnosed (#469). Thanks to mjcoynejr for testing the fix.
- BLASTing a mix of older V4 and newer V5 databases causes error. SequenceServer now catches and informs the user of this error. Thanks to Massimiliano babbucci for reporting this issue.
- On sub-URI deployments, clicking the back button would result in no databases to be displayed in the search form (#462). Fixed that. Thanks to Xiang Zhang for reporting the issue and testing the fix.
- The list of databases in the search form should be alphabetically sorted. This behaviour was lost in the rewrite leading to version 2.0 and has now been fixed. Thanks to Loraine Guéguen for reporting the issue.
- Docker-related optimisations: SequenceServer no longer prompts to join the mailing list when running under Docker (#468). Thanks to Matt for reporting the issue. Furthermore, we have updated the docker base image and reduced the image size by ~100 Mb.
- Documentation for HPC integration was out of date with regard to how to invoke the wrapper script (#445). Thanks to Loraine Guéguen for reporting the issue, testing the fix.
Thanks to Niek Art, Matt Yoder, Massimiliano babbucci, Bjoernsen, and Eric Y for reporting database compatibility issues that prompted us to create a path for migrating to the new database format.
2020-04-22 Release candidate 2
- A programming error prevented databases from being deselected when you go back to the search form after submitting a query. That’s now fixed. Thanks to Norman Johnson for reporting the issue.
- Database creation step could skip FASTA files containing too many Ns at the beginning of the file. Fixed that. Thanks to Michał T. Lorenc for reporting the issue.
- During the initial setup SequenceServer would print the message ‘config file not found’ multiple times. Fixed that.
- Results page would fail to load completely if the submitted queries resulted in no hits. Fixed that.
- Give users an option to sign up for release and other important announcements during the initial setup.
- Highlight the use of non-default parameters by changing the background of advanced parameters field to yellow when filled.
- Include SequenceServer version and a link to our paper in the search summary at the top of the results page.
New documentation
We have also been able to make many of the required changes to the website for the 2.0 release and are testing it locally: https://github.com/wurmlab/sequenceserver/tree/gh-pages2.0.
2020-03-15 New architecture for improved flexibility: 2.0 release candidate 1
Dear colleagues,
We are pleased to announce a candidate release of SequenceServer 2.0. This completes the rearchitecting of SequenceServer 1.0 that we announced almost two years ago: https://groups.google.com/d/msg/sequenceserver/2G4_jdAr4-k/2yxQKixVBQAJ
We have made many changes since version 1.0. The key features are:
- Support for BLAST 2.10.0 and the new BLAST database format.
- Results now open in a new page. You can share the link with colleagues or bookmark the page and return to it at a later date. Results are saved for 30 days.
- We have integrated three new visualisations that facilitate interpretation of BLAST results: Length distribution (histogram) of all hits of a query. This is inspired from our work with GeneValidator (https://academic.oup.com/bioinformatics/article/32/10/1559/1742817) and can, for example, help quality-assessment of gene predictions. Kablammo visualisation (https://academic.oup.com/bioinformatics/article/31/8/1305/212772) for each query-hit pair indicates clearly which parts of the query match which part of the hits. Circos-style plot of queries and their top hits. This is similar to the circoletto tool (https://academic.oup.com/bioinformatics/article/26/20/2620/194655). For example, it can help visually testing for conserved synteny or gene duplication.
- We provide a command-line mechanism for importing a BLAST XML file and visualising it in SequenceServer. This works for DIAMOND as well.
- The 30 hit limit for FASTA download has been removed. In addition, you can now download raw pairwise alignments.
- Better support for long-running BLAST searches, for rendering large BLAST results (thousands of hits), and for integrating as part of other websites.
- Improved error handling, security enhancements, and a new look!
Compared to the beta releases, the biggest remaining limitations were SequenceServer’s handling of large BLAST results and back button of browsers. We have addressed those.
We request you to try out the “candidate” release of version 2.0 and report any issues you encounter with your setup (or give us a thumbs up). We will update the documentation at http://sequenceserver.com over the next few weeks. In the meantime, please feel free to ask any question here or on GitHub.
Upgrading is simple. If you have Ruby 2.3 or later, run:
gem install --pre sequenceserver
This does not remove your current sequenceserver installation. Instead, it changes the default. If something breaks, you can rollback by running:
gem uninstall sequenceserver
Please report any issues here: https://github.com/wurmlab/sequenceserver/issues.
You can also send us pull-requests. We welcome any help you may be able to provide in terms of improving test coverage and making code style checkers happy.
Kind regards, Priyam & Yannick
2019-12-21 Announcing 2.0.0beta4
Dear all,
We are pleased to announce the last beta release of this series. In this release, we have added a few features based on popular demand and fixed tons of bugs.
Overall, the new architecture now feels quite robust and has seen a growing adoption within the community. We have incremented the version number to 2.0 to reflect this milestone. We thank you for your patience, encouragement, and support.
New features since 1.1.0-beta12
- Update to BLAST+ 2.9.0
- Ability to import BLAST results in XML format from DIAMOND and BLAST versions 2.2.30 - 2.9.0
- Ability to pre-populate text area with server-sent query sequences (thanks to Richard Challis for feature suggestion and initial implementation)
- On startup, sequenceserver now tries to determine the ip and the hostname of the server and prints two URLs that may be suitable for sharing with colleagues
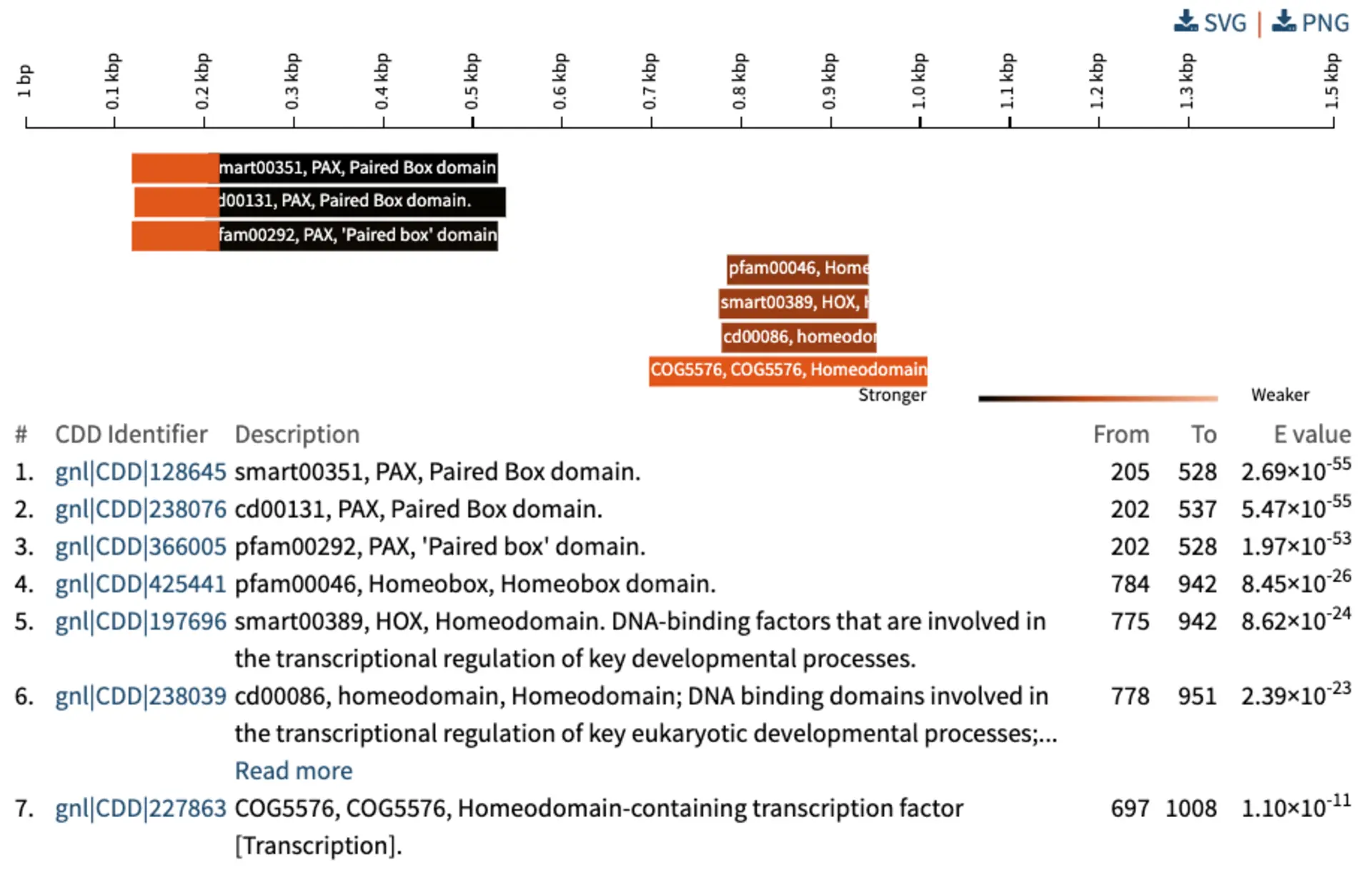
- Recognise PFAM and RFAM hit/subject ids and include a link to the hits PFAM or RFAM page (credit: Tomas Pluskal)
Bug fixes and refinements
- In a suburi deployment (e.g., antgenomes.org/blast), take suburi into account when constructing url for the results page (thanks to Guy Leonard for reporting the issue)
- Use more specific regex for determining non -parse_seqids databases. Otherwise, presence of
gn|in sequence ids would disable sequence retrieval (thanks to Josh Goodman for reporting the issue) - Length of query and hit sequences are now easily accessible in the report page. This is included in the query and hit header, on the right hand side, in a gray font
- Restored the ability to fold and un-fold individual hits (credit: Josh Goodman)
- Removed .00 from decimal numbers (i.e., 100.00% is now shown as 100%)
- Fixed vertical alignment of links and buttons provided for each hit
- Removed trailing comma in HSP statistics line for BLASTP
- Properly wrap text in the summary overview on top
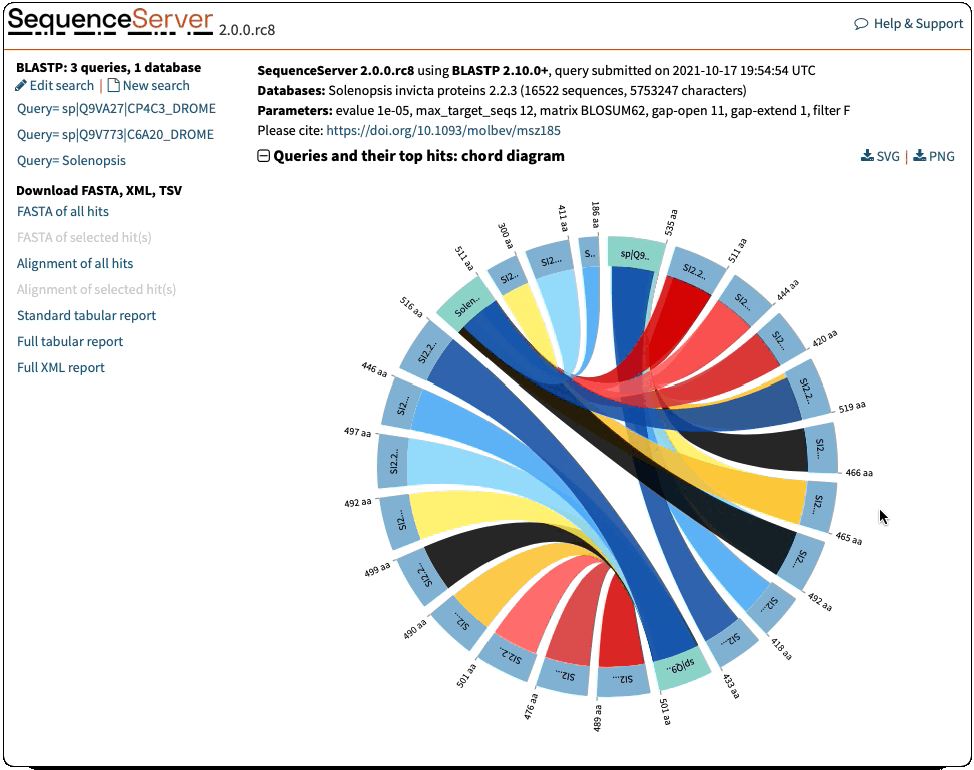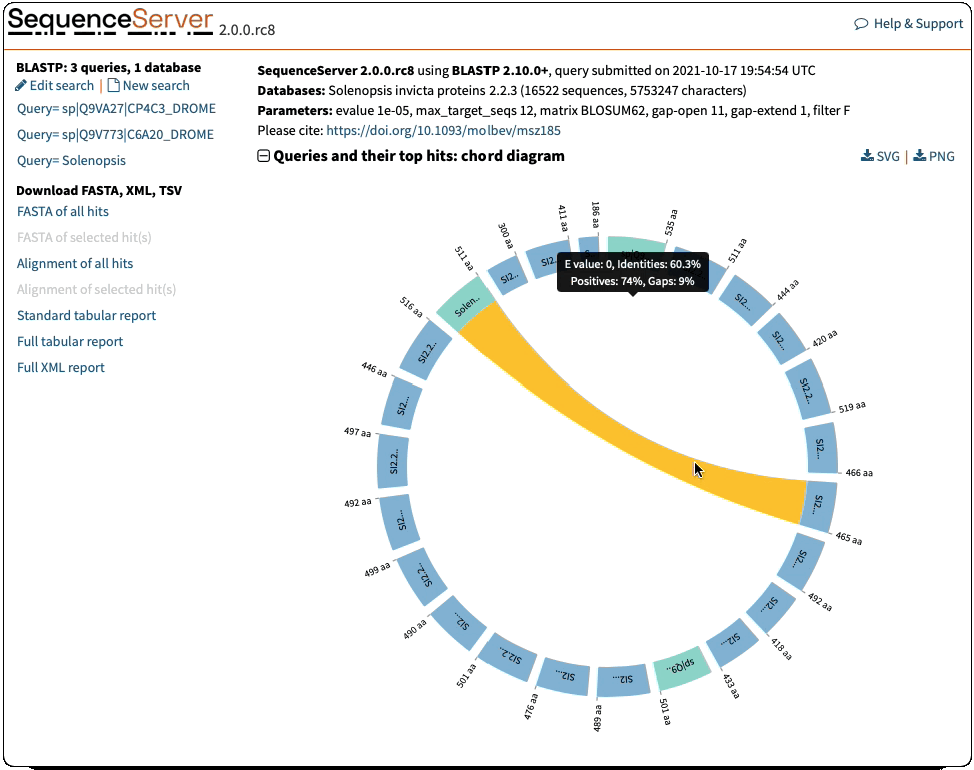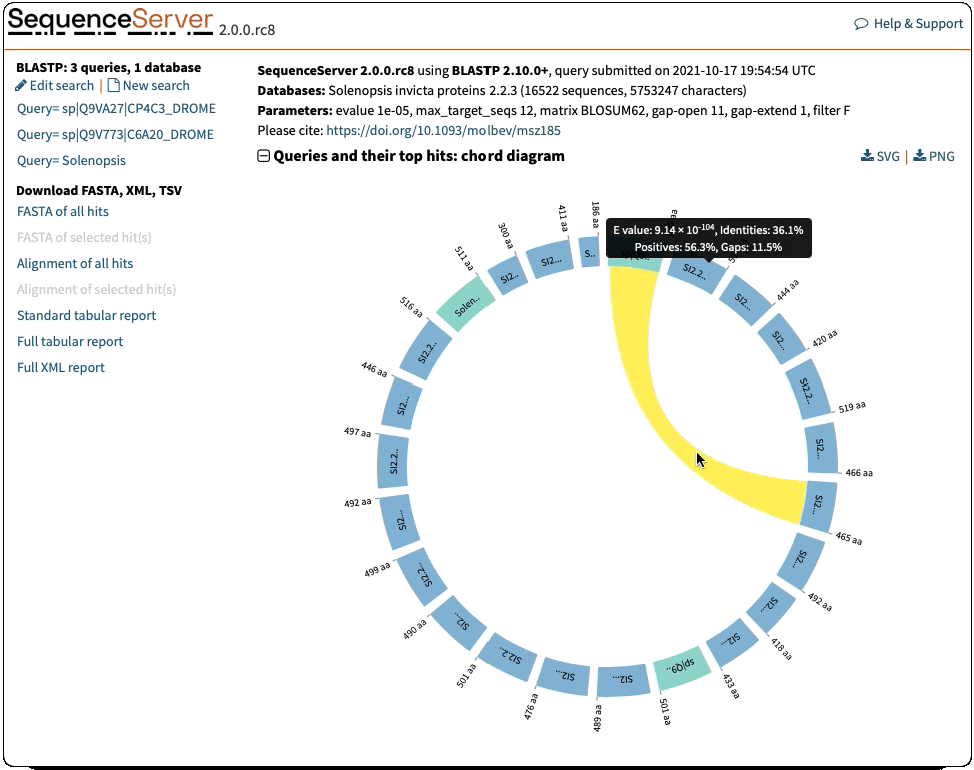
Circos visualisation:
- Sequence lengths were erroneously downscaled by 100. Fixed that
- Formatting of e-value and identities in the tooltips was inconsistent with the rest of the report. Fixed that
- Ribbons had no colour for the specific case of 1 query and 2 hits. Fixed that
- Filter out queries with no hits before drawing the visualisation
- Change to more meaningful title ‘Chord diagram: queries and their top hits’
Length distribution of all hits for each query:
- When query and database are in different sequence space (DNA or protein), normalise query length to match database space (#409)
- Reduced the number of ticks in the y-axis otherwise the ticks looked too “squeezed up” when there were too many hits
- Round up the number of ticks on x-axis so that the scale ends consistently in a major or a minor tick
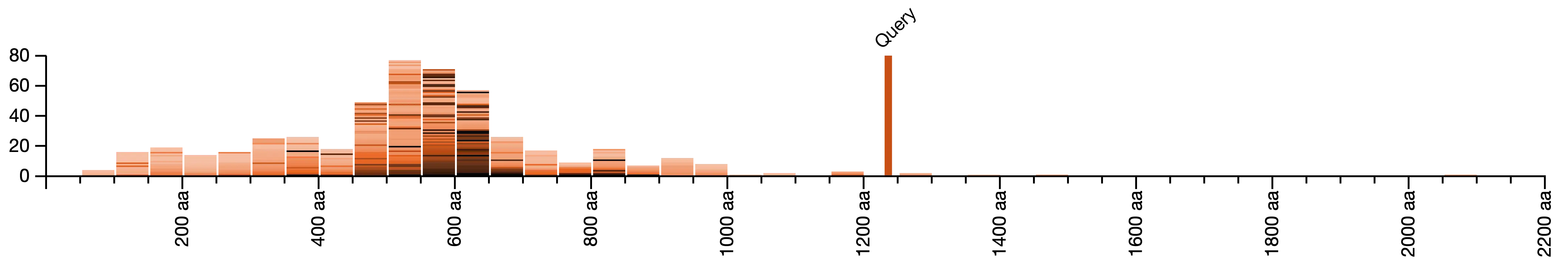
Code quality improvements
- Fixed many of the warning messages generated by React in browser console
- Fixed style guide violations in both Ruby and JavaScript code
- Expanded test coverage and improved automated testing
2018-12-20 Announcing the last release of the SequenceServer beta series, and version 2.0
Dear all,
We are pleased to announce the last beta release of this series. In this release, we have added a few features based on popular demand and fixed tons of bugs.
You can find a detailed list of changes on our GitHub release page: https://github.com/wurmlab/sequenceserver/releases/tag/2.0.0.beta4.
Overall, the new architecture now feels quite robust and has seen a growing adoption within the community. We have incremented the version number to 2.0 to reflect this milestone. We thank you for your patience, encouragement, and support.
We invite you to try it out and help us get to stable version by sending us a pull-request or reporting any bugs on our issue tracker: https://github.com/wurmlab/sequenceserver/issues
Upgrading is simple. If you have Ruby 2.3 or later, run:
gem install --pre sequenceserver
This does not remove your current sequenceserver installation. Instead, it changes the default. If something breaks, you can rollback by running:
gem uninstall sequenceserver
We welcome any help you may be able to provide in terms of code review, improving test coverage, and making code style checkers happy.
2018-09-10 Announcing a 12th Beta release of SequenceServer 1.1.0 for testing
Hello,
We are pleased to announce 12th beta release of SequenceServer 1.1.0. This release includes a few small enhancements and several code-style improvements.
Changelog
- Write default BLAST options that were previously defined only in the code to config file during setup. Since old config files won’t have this info available, retain the old behaviour of automatically adding ‘-task blastn’ to BLASTN searches and log this during startup. (issue raised by Niek Art)
- Multipart database volumes can have 3 digit suffixes (e.g. refseq_genomic.100). Our regex allowed for only 2 digit suffixes (e.g. nr.00). Fixed that. (issue raised by Niek Art)
- Show location of config file by default during startup
- Code-style improvements. (credit: Emeline Favreau, Esteban Gomez)
We invite you to try out the beta releases and help us get to stable version by sending us a pull-request or reporting any bugs on our issue tracker: https://github.com/wurmlab/sequenceserver/issues
Upgrading is simple. If you have Ruby 2.3 or later, run:
gem install --pre sequenceserver
This does not remove your current sequenceserver installation. Instead, it changes the default. If something breaks, you can rollback by running:
gem uninstall sequenceserver
We welcome any help you may be able to provide in terms of code review, improving test coverage, and making code style checkers happy.
2018-07-10 Announcing a 11th Beta release of SequenceServer 1.1.0 for testing
Hello,
We are pleased to announce 11th beta release of SequenceServer 1.1.0. This release includes many small-small bug fixes and enhancements.
Changelog
- Display of large results are handled by rendering results of fifty queries at a time. However, in each cycle of update, instead of focusing only on the next fifty queries, React would automatically also check if previously rendered queries needed to be updated as well. This resulted in each cycle of update to take more and more time. Prevent that by returning false from shouldComponentUpdate lifecycle method if the query is already rendered.
- Kablammo / graphical HSP overview:
- Copy-pasting hit sequence from sequence viewer should eliminate whitespace introduced by grouping of residues, but it didn’t. So disabled grouping of residues in sequence viewer so that the sequence can be copy-pasted as it is.
- SVG and PNG download links would break into two lines on smaller windows. Fixed that.
- Do not change padding when hit is selected. Further, apply orange border to selected hits only on the left side instead of on all sides.
- Downloading alignments of selected hits wouldn’t work if sequence ids included a pipe character. Fixed this.
- Last line of the alignment would sometimes not be shown. Fixed that.
- Expanded error handling:
- Inform user if BLAST’s XML output is invalid, and suggest deduplicating sequence ids or rerunning with a single database (issue #194) (credits: Ben Woodcroft, Tomáš Pluskal, and others).
- Inform user if BLAST ran out of memory and ask to retry with smaller query. Should be rare, but is good to know when this happens.
- Inform user if SequenceServer doesn’t have permission to create job directory. Should be particularly helpful when setting up with Apache.
- Inform user if SequenceServer/BLAST runs out of disk space.
- Inform user if BLAST produced empty output.
- Update Dockerfile so that docker image is built from the source. This ensures that the tag applied to docker image and software version are in sync. (credit: Michał T. Lorenc)
- Don’t use symlinks inside of the repository as symlinks are not supported on all platform and can thus cause installation to fail.
- Ensure the new version can be deployed to a suburi.
Roadmap
https://docs.google.com/document/d/1Vt2MmaD5h5oN8XrmokLVnjUfRLWqKVWeEja3w9SIeBw (comments welcome)
We invite you to try out the beta releases and help us get to stable version by sending us a pull-request or reporting any bugs on our issue tracker: https://github.com/wurmlab/sequenceserver/issues
Upgrading is simple. If you have Ruby 2.3 or later, run:
gem install --pre sequenceserver
This does not remove your current sequenceserver installation. Instead, it changes the default. If something breaks, you can rollback by running:
gem uninstall sequenceserver
We welcome any help you may be able to provide in terms of code review, improving test coverage, and making code style checkers happy.
2018-06-19 Announcing a 10th Beta release of SequenceServer 1.1.0 for testing
Hello,
We are pleased to announce 10th beta release of SequenceServer 1.1.0. Based on popular demand we added a button to select all databases. We have further made several visual and speed improvements to the results page and fixed an important bug.
Features
- Added a button to select all databases.
- Meta data on top of report page now includes date (as well as labels for program, databases, parameters).
- Reduce overall amount of vertical space on the report page so that more results fit in the same height.
- Detailed space reduction effort were needed for pairwise alignments, which now use almost 2x less vertical space. Importantly, alignments are now “pretty formatted” in the browser instead of on the server so that all available horizontal space is known beforehand. Previously we were restricted to assuming 60 characters per line.
- For displaying pairwise alignments we previously used nested tables. We now use list of divs and pre tags. This reduces memory usage and increases speed (crucial for large reports).
- Eliminate borders from around graphs to reduce clutter and free up vertical space.
- Add % sign for when identity, gaps, positive are shown as percentage.
- Use more meaningful headers for hits and hsp overview graphs.
Bug fixes
- Consecutive blank spaces in the middle line of the alignment were truncated while parsing BLAST’s XML output. Fix it.
Roadmap
https://docs.google.com/document/d/1Vt2MmaD5h5oN8XrmokLVnjUfRLWqKVWeEja3w9SIeBw (comments welcome)
We invite you to try out the beta releases and help us get to stable version by sending us a pull-request or reporting any bugs on our issue tracker: https://github.com/wurmlab/sequenceserver/issues
Upgrading is simple. If you have Ruby 2.3 or later, run:
gem install --pre sequenceserver
This does not remove your current sequenceserver installation. Instead, it changes the default. If something breaks, you can rollback by running:
gem uninstall sequenceserver
We welcome any help you may be able to provide in terms of code review, improving test coverage, and making code style checkers happy.
2018-05-30 Announcing a 8th Beta release of SequenceServer 1.1.0 for testing
Hello,
We are pleased to announce 8th beta release of SequenceServer 1.1.0. This is a small release that fixes an issue introduced by the previous release.
Changelog
- Fix programming errors in Report.fetchResults so that it behaves as intended (poll with decreasing frequency and updates page when results are available). Reported by Devon Ryan.
- Change title of hits overview visualisation to matching sequences.
We invite you to try out the beta releases and help us get to stable version by sending us a pull-request or reporting any bugs on our issue tracker: https://github.com/wurmlab/sequenceserver/issues
Upgrading is simple. If you have Ruby 2.3 or later, run:
gem install --pre sequenceserver
This does not remove your current sequenceserver installation. Instead, it changes the default. If something breaks, you can rollback by running:
gem uninstall sequenceserver
We welcome any help you may be able to provide in terms of code review, improving test coverage, and making code style checkers happy.
Roadmap: https://docs.google.com/document/d/1Vt2MmaD5h5oN8XrmokLVnjUfRLWqKVWeEja3w9SIeBw (comments welcome)
2018-05-26 Announcing a 7th Beta release of SequenceServer 1.1.0 for testing
Hello,
We are pleased to announce 7th beta release of SequenceServer 1.1.0. In this release we have addressed inconsistencies created by asynchronous rendering of results that was introduced in previous release, made a few enhancements and bug fixes, and introduced one final visualisation.
Changelog
- Add Kablammo (http://kablammo.wasmuthlab.org) to visualise HSPs per hit (credit: Alekhya Munagala).
- Ensure all scripting of results HTML (such as binding event handlers) is done at appropriate stages of rendering process.
- Report page repeatedly pings the server for results. The frequency of pings was meant to decrease with time. However, a bug in the code resulted in frequency of pinging to remain the same. Fix that.
- When there are too many hits to create circos visualisation, show a message explaining the same (credit: Hiten Chowdhary).
- Do not add circos visualisation to page if there are no hits to visualise (credit: Hiten Chowdhary).
What’s next
The idea is to continue with optimising results page and individual visualisations. Apart from optimisations, only a few more small-small features remain to be implemented before the final release.
Roadmap: https://docs.google.com/document/d/1Vt2MmaD5h5oN8XrmokLVnjUfRLWqKVWeEja3w9SIeBw (comments welcome)
We invite you to try out this release and help us get to a stable version by sending us a pull-request or reporting any bugs on our issue tracker: https://github.com/wurmlab/sequenceserver/issues
Upgrading is simple. If you have Ruby 2.3 or later, run:
gem install --pre sequenceserver
This does not remove your current sequenceserver installation. Instead, it changes the default. If something breaks, you can rollback by running:
gem uninstall sequenceserver
We welcome any help you may be able to provide in terms of code review, improving test coverage, and making code style checkers happy.
2018-05-21 Announcing a 6th Beta release of SequenceServer 1.1.0 for testing
Hello,
We are pleased to announce 6th beta release of SequenceServer 1.1.0 for testing. In this release we have enabled one more visualisation, focused on optimising the results page, and fixed some bugs.
Changelog
- Show length distribution of hits per query (credit: Hiten Chowdhary). Like circos, this visualisation is drawn only when user clicks the corresponding button.
- Do not draw alignments overview (per query) on page load if number of queries is >250. In this case, the user must click the corresponding button to render the visualisation.
- Once all results have been retrieved from the server, update the page with
results of 50 queries every 500 ms. Along with the optimisation above, this
allows us to render large results - tested up to 1200 queries and 1500 hits.
The page remains responsive (e.g., scrolling) while it is being updated and
after, however, there are two caveats still when viewing large results (we
plan to address these in the next beta release):
- Some features, such as selecting hits for download, ability to download FASTA and pairwise alignments for a single hit, and affixing of sidebar are not activated till the entire result has been rendered and currently there is no clear way to communicate this to the user.
- Visualisations are slow to render and hide on click.
- Displaying of appropriate error messages when BLAST failed was broken due to a typo in the code. Fixed it (credit: Tomáš Pluskal).
- Make entering taxid optional again when creating BLAST databases from FASTA file (reported by: Tomáš Pluskal)
What’s next (comments welcome)
https://docs.google.com/document/d/1Vt2MmaD5h5oN8XrmokLVnjUfRLWqKVWeEja3w9SIeBw
We invite you to try out this release and help us get to a stable version by sending us a pull-request or reporting any bugs on our issue tracker: https://github.com/wurmlab/sequenceserver/issues
Upgrading is simple. If you have Ruby 2.3 or later, run:
gem install --pre sequenceserver
This does not remove your current sequenceserver installation. Instead, it changes the default. If something breaks, you can rollback by running:
gem uninstall sequenceserver
We welcome any help you may be able to provide in terms of code review, improving test coverage, and making code style checkers happy.
2018-04-27 Announcing a 5th Beta release of SequenceServer 1.1.0 for testing
Hello,
We are pleased to announce 5th beta release of SequenceServer 1.1.0. This release activates the circos visualisation for testing and fixes a few other bugs. Detailed changelog and a rough roadmap to stable release below.
Changelog
- Enable Circos visualisation (experimental). The visualisation is drawn specifically on clicking the ‘CIRCOS’ link on the report page. Works best when total number of matching hits and hsps is small.
- Entering non-numeric taxid when creating BLAST databases would cause SequenceServer to crash. Instead, it now prompts user to re-input taxid.
- Anchor tags as well as buttons are used on the report page for different actions. Ensure both are consistently styled.
- We use SauceLabs browser testing platform for testing the user-interface. Duly credit SauceLabs in README by linking to their website.
Roadmap (comments welcome)
https://docs.google.com/document/d/1Vt2MmaD5h5oN8XrmokLVnjUfRLWqKVWeEja3w9SIeBw
We invite you to try out this release and help us get to a stable version by sending us a pull-rquest or reporting any bugs on our issue tracker: https://github.com/wurmlab/sequenceserver/issues
Upgrading is simple. If you have Ruby 2.3 or later, run:
gem install --pre sequenceserver
This does not remove your current sequenceserver installation. Instead, it changes the default. If something breaks, you can rollback by running:
gem uninstall sequenceserver
We welcome any help you may be able to provide in terms of code review, improving test coverage, and making code style checkers happy.
2018-04-20 Announcing a 4th Beta release of SequenceServer 1.1.0 for testing
Hello,
We are pleased to announce 4th beta release of SequenceServer 1.1.0 for testing.
Changelog
- Retain the order in which hits and hsps are returned by BLAST. In previous beta releases, hits and hsps were sorted by e-value, which didn’t work well when multiple e-values were similar.
- Show error message (SystemError) if no permission to create job directory, or no disk space.
- By default, delete finished jobs after 30 days instead of 7 days.
- Ensure browser cache is refreshed for new releases.
- Do not show backtrace when job not found.
- Refactoring in previous beta release had broken XML and tabular report download link on the results page - fix it.
- Slightly modify citation line.
We invite you to try out this release and help us get to a stable version by reporting any bugs on our issue tracker (https://github.com/wurmlab/sequenceserver/issues) or sending us a pull-request.
Upgrading is simple. If you have Ruby 2.3 or later, run:
gem install --pre sequenceserver
This does not remove your current sequenceserver installation. Instead, it changes the default. If something breaks, you can rollback by running:
gem uninstall sequenceserver
We welcome any help you may be able to provide in terms of code review, improving test coverage, and making code style checkers happy.
2018-04-12 Announcing a 3rd Beta release of SequenceServer 1.1.0 for testing
Hello,
We are pleased to announce 3rd beta release of SequenceServer 1.1.0 for testing.
Changelog
- Expanded User Interface (UI) testing to cover more scenarios:
- All five basic BLAST algorithms are now tested using a multi-fasta query with no defline for the first sequence, and at least two databases. UI tests previously covered selected BLAST algorithms and were restricted to single query sequence and single database.
- Expanded platforms on which automated UI tests are run (powered by:
SauceLabs) -
- Latest Chrome and Firefox on Linux, macOS 10.13, and Windows 10
- Latest Safari on macOS 10.13
- Latest Edge on Windows 10 Previously we tested only on latest Firefox on Linux.
- Update error handling mechanism to work with the new asynchronous nature of running BLAST searches (credit: Hiten Chowdhary).
- Improve error reporting:
- Fix parsing of error message when BLAST exit status is 1 (incorrect advanced params or weird error in query sequence)
- BLAST results are deleted after 7 days. If user attempts to access a deleted job, show ‘job not found’ message instead of treating it like other errors.
For those interested in using SequenceServer API to run BLAST searches using curl or so,
- Errors are now reported in JSON format as well instead of HTML (error body may still contain HTML links)
We invite you to try out this release and help us get to a stable version by reporting any bugs on our issue tracker (https://github.com/wurmlab/sequenceserver/issues) or sending us a pull-request. Some standing issues:
- The expanded UI testing has revealed problems with Chrome 48 on Linux and latest Edge 16 on Windows 10 (other failures in the browser matrix in README are from temporary network issues)
Upgrading is simple. If you have Ruby 2.3 or later, run:
gem install --pre sequenceserver
This does not remove your current sequenceserver installation. Instead, it changes the default. If something breaks, you can rollback by running:
gem uninstall sequenceserver
We welcome any help you may be able to provide in terms of code review, improving test coverage, and making code style checkers happy.
2018-03-30 Announcing a 2nd Beta release of SequenceServer 1.1.0 for testing
Hello,
We are pleased to announce 2nd beta release of SequenceServer 1.1.0 for testing.
Changelog
- Addressed all the failing tests - all tests now pass! Thanks to Hiroyuki Nakamura for fixing Travis test script.
- Show the clickable list of query ids in the sidebar for only up to 8 queries, so that the sidebar doesn’t spill over onto the footer.
- Update to Sinatra 2.0, so that SequenceServer can be embedded in Rails 5 (credit Hiroyuki Nakamura).
- Speed up page load time by compressing server’s response on the go. Both the search form and short BLAST results now load significantly faster.
We invite you to try out this release and help us get to a stable version by reporting any bugs on our issue tracker (https://github.com/wurmlab/sequenceserver/issues) or sending us a pull-request.
Upgrading is simple. If you have Ruby 2.3 or later, run:
gem install --pre sequenceserver
This does not remove your current sequenceserver installation. Instead, it changes the default. If something breaks, you can rollback by running:
gem uninstall sequenceserver
We welcome any help you may be able to provide in terms of code review, improving test coverage, and making code style checkers happy.
2018-03-22 Announcing a major new version of Sequenceserver 1.1.0-beta
Dear all,
We are pleased to announce a “beta” release of the next major version of SequenceServer - 1.1.0-beta for testing.
We have fundamentally restructured how SequenceServer works in order to achieve two goals.
- Most importantly, with the approach used to date, we were limited in what we could do with visualisations and filtering. The new approach overcomes all of this.
- We felt it was important to be able to bookmark and share BLAST results, and be able to return to them at a later date.
This new version incorporates multiple new minor features (full list below for details), but the biggest change is the “under the hood” restructuring. Today’s beta release is experimental - it has some limitations we know about, and we hope that you can help us find those that remain unknown. We expect this release to be the first of a series of small regular improvements each incorporating additional new features.
While we have begun using this version for real things, we consider it a “development” rather than a “production” release. Please use with care and report any problems on the github tracker or on this thread.
Features
- Update to BLAST 2.6.0+
- We put a customizable default parameter in BLAST searches to only return hits with evalues stronger than 1e-5 (BLAST’s default of e-value 10 includes too many spurious hits that are confusing to junior users)
- We make it explicit that ‘-task blastn’ option is used for BLASTN (instead of BLASTN’s default behaviour of running megablast)
- Stable URL for results: meaning that results page can now be bookmarked, shared, or linked to from your lab notebook
- “Download FASTA of all” link in the sidebar can handle many sequences (previously it was disabled if >30 hits)
- Ability to download individual or all alignments in FASTA format (code from Kablammo project at http://kablammo.wasmuthlab.org/)
- Ability to download alignment overview in SVG and PNG image (code from Kablammo project at http://kablammo.wasmuthlab.org/)
- Use a, b, c numbering for HSPs to distinguish easily from hits
- Show summary of BLAST database and search parameters at top of results page
- Show coverage and identity in hits table
- results can be opened in a new tab by pressing ‘Cmd’ / ‘Ctrl’ button while clicking on the submit button (like any other link).
- Drop support for Ruby older than 2.3. This allows SequenceServer to be embedded in Rails 5, means we benefit from performance improvements in new Ruby, and importantly makes code and dependencies easier to maintain and test
- If BLAST was downloaded by SequenceServer or path to BLAST binaries was explicitly provided by the user, BLAST searches are run in “sandbox mode”. In sandbox mode, SequenceServer cannot access any other system command. This increases security.
- We no longer require BLAST databases to have been created with -parse_seqids option of makeblastdb
Bugfixes
- Alignment overview: colour each HSP by its evalue (instead using the e-value of the hit)
- Fix crash when downloading amino acid sequences that include stop codons (*)
- All code and fonts (except Twitter badge) are loaded locally
Known issues
As this is beta software, a few things may not work as expected. Following is a list of known issues that we are currently working on:
- Performance with many number of hits: e.g., sequenceserver doesnt load the results of BLASTing a 300 kilobase file including >1000 queries against itself fails.
- Deploying on suburi (e.g., antgenomes.org/sequenceserver) currently doesn’t work
- Hits table can look jumbled up if BLAST search returns multiple hits with same evalue
- Sidebar height is not appropriately constrained
- 6 tests fail on the Travis code checker
Technical explanation of changes
- We now use browser’s default form submission behavior instead of using AJAX. This allows results to be opened in a new tab by pressing ‘Cmd’ / ‘Ctrl’ button while clicking on the submit button (like any other link).
- Previously, submitting the search form and obtaining results from the server would be accomplished in one long HTTP request. This was problematic for long running BLAST searches as production servers (e.g., Nginx, Apache) drop connections after a set interval. Now search form submission creates a ‘job’ (with a unique identifier) whose status is queried repeatedly by the result page. Such ‘asynchronous’ approach is more scalable and should also work well with job schedulers like qsub and bsub.
- The server now returns results in JSON format instead of HTML format. We use the React framework along with JSX to generate HTML results directly in the browser. Access to raw results directly in the browser has enabled us to test a few different visualisations for BLAST results (not part of this release). This approach also allows the use of ‘curl’ to obtain results in command line and process the results in any scripting language.
We invite you to try out this release and help us get to a stable version by reporting any bugs on our issue tracker (https://github.com/wurmlab/sequenceserver/issues) or sending us a pull-request.
Upgrading is simple. If you have Ruby 2.3 or later, run:
gem install --pre sequenceserver
This does not remove your current sequenceserver installation. Instead, it changes the default. If something breaks, you can rollback by running:
gem uninstall sequenceserver
We welcome any help you may be able to provide in terms of code review, improving test coverage, and making code style checkers happy.
Kind regards, Priyam & Yannick
https://wurmlab.com Evolutionary genomics @ Queen Mary U London