Search Raw Reads from NCBI’s SRA Database
We are thrilled to unveil an important addition to SequenceServer Cloud.
Our newest feature enables direct searching of raw reads from the NCBI’s Sequence Read Archive (SRA) database. This means you can search the stupendously large amounts of raw Illumina, Nanopore, Pacbio reads from each sample in every genomic and RNA-seq experiment. And also the smaller amounts from old Solexa and 454 runs.
This ability is critical for research questions that cannot be simply answered by examining assembled transcriptomes, genomes, or metagenomes, or by seeing summary counts of mapped reads.
How It Works
As outlined in the video above, it’s straightforward.
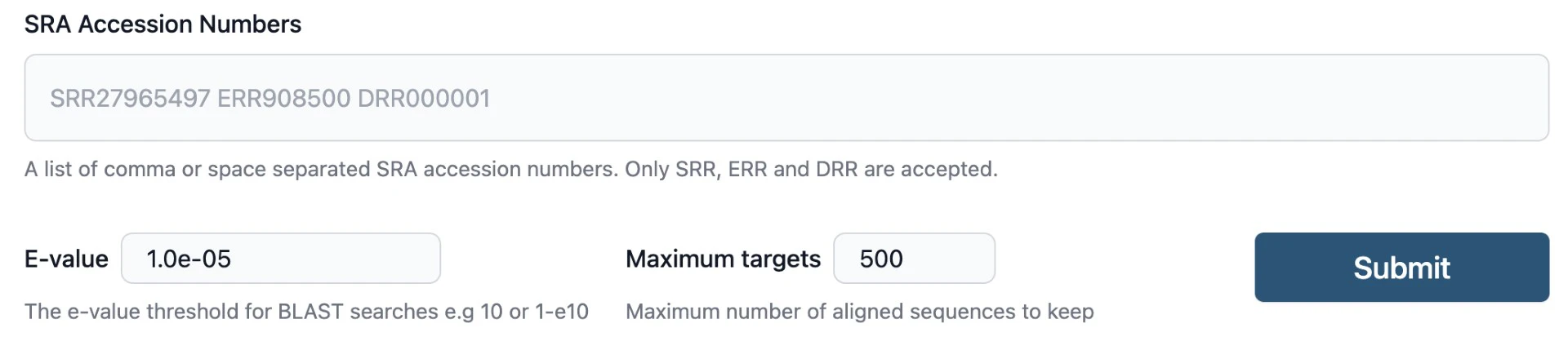
- Input Your Query: Paste your FASTA query sequence(s) into the search field.
- Select Your SRA Runs: Specify the identifiers of the SRA runs of interest.
- Specify RNA Analysis (If Applicable): For RNA-related searches, click “Allow spliced alignments”, so splicing isn’t penalized.
- Specify Parameters: The SRA BLAST user interface allows for setting E-values and the maximum number of aligned sequences to keep when submitting a query.
- Submit: Hit Submit. Each SRA is analyzed in parallel. This takes minutes to hours depending on data sizes involved (some SRA datasets are really big!)
- View Results: (see below)
Making Sense of SRA BLAST Analysis Results
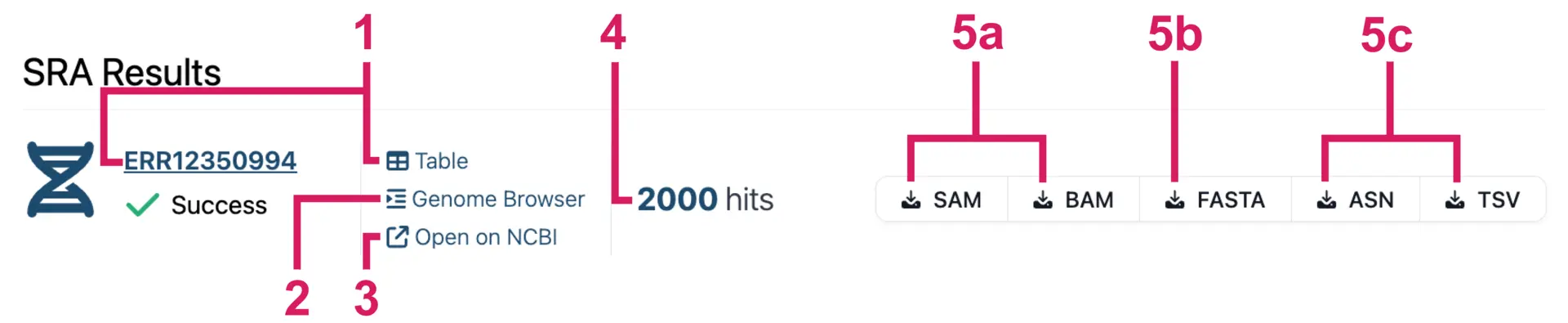
Results appear progressively as the analysis of each SRA run finishes. For each SRA sample, you get:
- A results table of all the alignments. You can view hits in a table by clicking on the accession number or the “Table” button.
- An interactive “Genome Browser” to visualize alignments.
- “Open on NCBI” goes to the web page for the SRA accession.
- The number of reads that match/hit the query sequence.
- Export options for the alignment reads.
- a. Download alignments in SAM/BAM format, which includes CIGAR strings and MD tags that indicate mismatches and indels.
- b. Download all the hit sequences in FASTA format.
- c. Download ASN and TSV reports that contain BLAST alignment statistics.
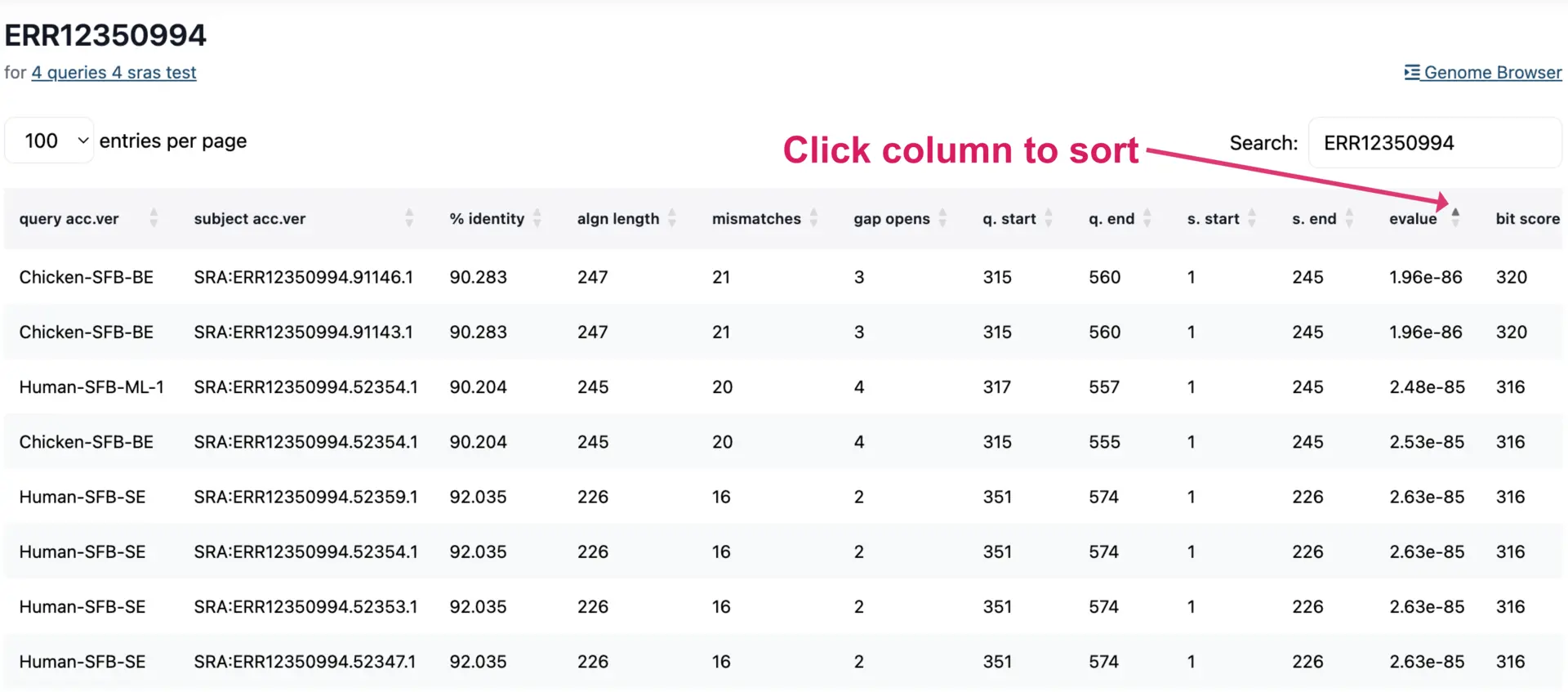
Viewing Results in Table Format
The results table can be viewed interactively or downloaded as a TSV file. The table contains lots of information, such as E-values, percentage identity, and other attributes for each hit. Each column can be sorted to view and prioritize alignment metrics.
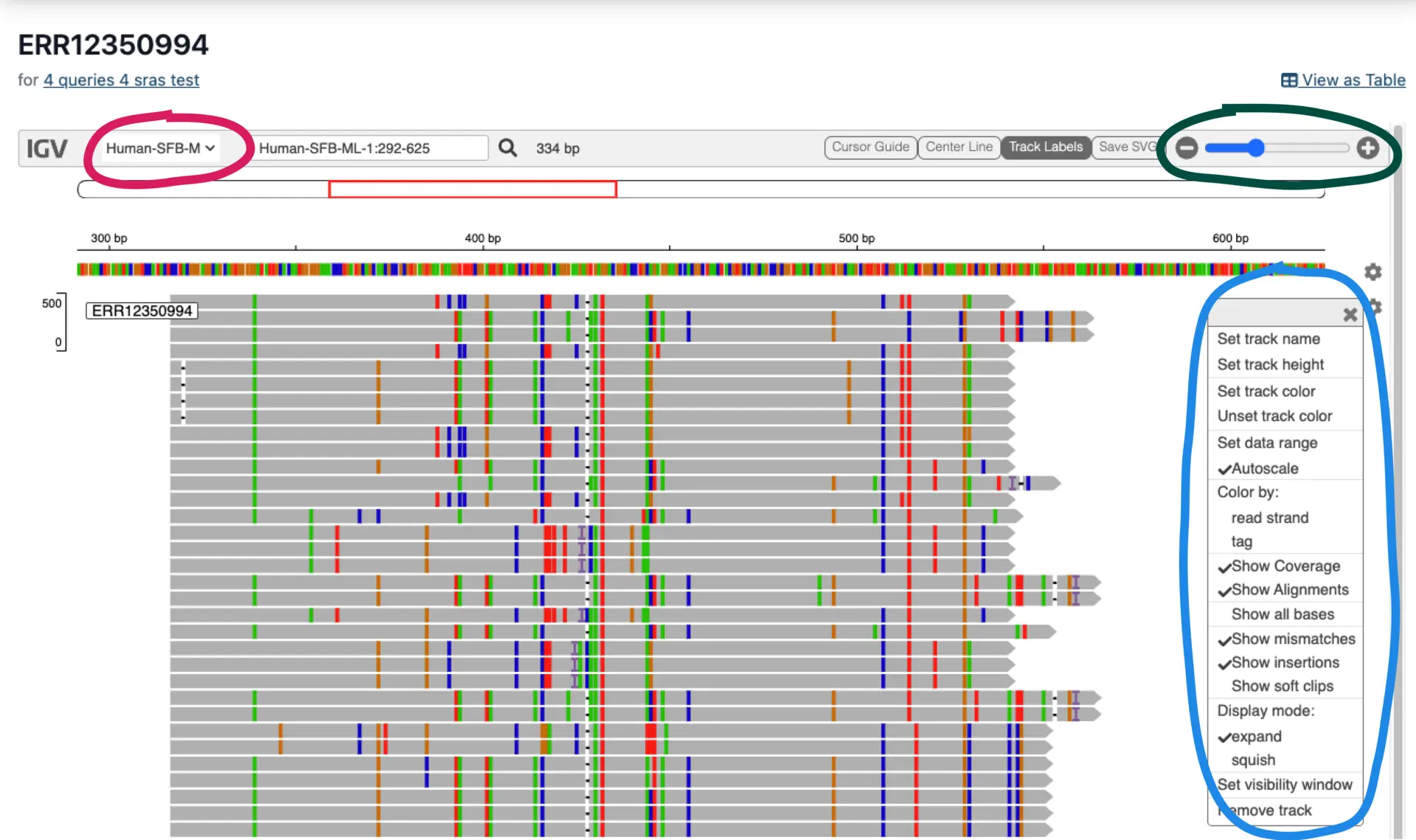
Viewing Results in the Interactive Genome Browser
The interactive genome browser enables users to view alignments for each SRA accession. Alignment attributes like SNPs, INDELs, and read orientation options can be toggled (blue circle) and explored at different magnifications (green circle). Each query sequence can be selected at the top left (red circle).
Potential Applications of SRA BLAST
We’re excited to see how SequenceServer Cloud users will use SRA BLAST. Likely applications include:
- Exploring viral sequences to decipher patterns of evolution and spread across species and over time.
- Examining transposable element activity to understand how they jump, evolve, and shape phenotypes.
- Environmental Genomics: Analyzing microbial communities from soil microbiomes to the human gut, for example to understand where particular genes and accessory genomes are found, and in which frequencies.
Happy BLASTing!


