Identifying conserved protein domains when running SequenceServer Cloud BLAST searches
SequenceServer is great for quickly and securely BLASTing unpublished datasets. We now also identify conserved protein domains for every query sequence. For this, we use the NCBI CDD conserved domain database. Results are displayed as part of the BLAST report, and can also be downloaded for further processing.
CDD screenshots
These screenshots show what this looks like; we provide more context below.
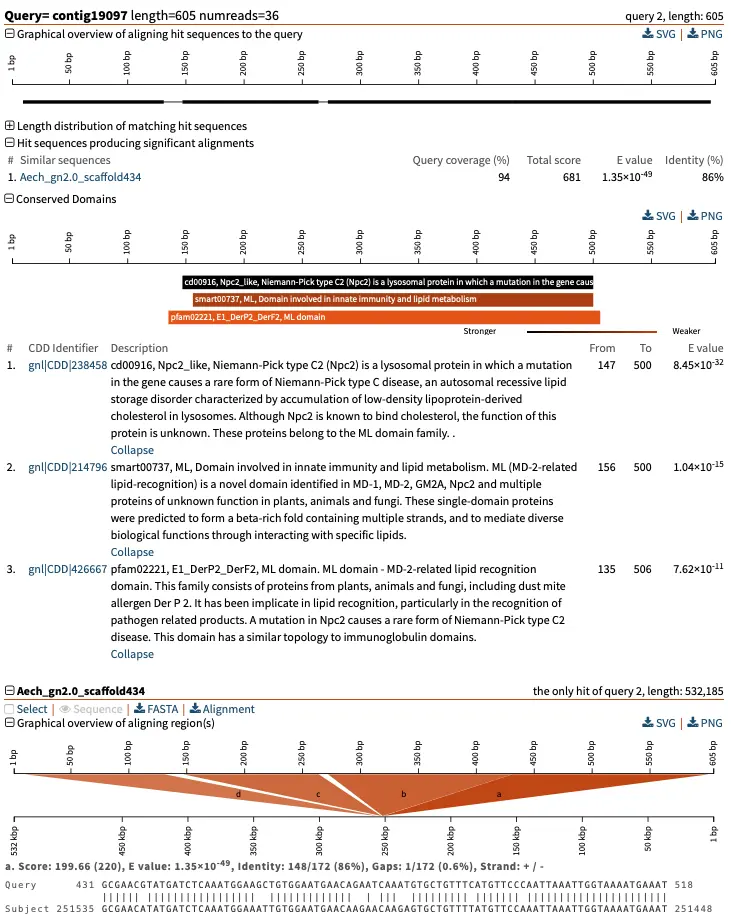
Example of endonuclease domain identification
For each query, we end up with an additional plot, like this one, which shows that the query sequence contains a conserved endonuclease domain. Here, the standard CDD database includes several versions of that domain that differ slightly, which is why we see several aligning domain models.
Example of other output data
A standard SequenceServer BLAST report now also includes a table with:
- the domain models that were identified,
- E-values indicating the strength of the matches,
- and the coordinates of where the query sequence matches the domain model.
Raw data download
Domain identification results are also available as a separate download in JSON format. This is useful if you want to do further analysis on large numbers of genes, or for general functional protein annotation.
Why protein domains?
The conserved domain database (CDD) is a collection of protein domains that are conserved across species. It is a great resource to identify the functions of a protein. Each domain model was built from a multiple alignment of many sequences.
Using a domain-identification method is complementary to a normal BLAST-type sequence similarity search: Sometimes you may get 0 BLAST hits, but a protein domain identification tool can still detect protein domains. This type of model can be more sensitive than aligning to individual sequences.
What is a protein domain?
A protein domain is a part of a protein that has a specific function. For example, a protein domain can be a binding site for a ligand, or a site that is involved in catalysis. Based on alignments of sections of proteins that have a similar function, protein domains can be identified.
That was done for example for the protein kinase domain. This domain is 250 amino acids long and found in proteins that are involved in phosphorylation.
Hundreds of other protein domains are known and included in NCBI’s CDD. Some classic examples:
- SH2 Domain (Src Homology 2 Domain): This domain binds to phosphorylated tyrosine residues and plays a critical role in signal transduction pathways. It’s often found in proteins involved in cell signaling and regulation.
- PH Domain (Pleckstrin Homology Domain): This domain is involved in targeting proteins to different membranes, where they bind to phosphoinositides. It’s crucial for a variety of cellular processes including cell signaling and membrane trafficking.
- Zinc Finger Domain: A small, functional, independently folded domain that coordinates one or more zinc ions to help stabilize its structure. It’s commonly found in proteins involved in DNA binding and gene regulation.
- HLH Domain (Helix-Loop-Helix Domain): This domain is involved in protein-protein interactions and is common in transcription factors. It’s characterized by two α-helices connected by a loop.
- Ig Domain (Immunoglobulin Domain): This is a type of domain that is commonly found in the immunoglobulin (antibody) family of proteins, as well as in many other types of proteins. It’s involved in protein-protein interactions and has a characteristic sandwich-like structure.
- HOX domain (Homeobox domain): This is a DNA-binding domain found in homeobox genes. It regulates developmental processes by controlling gene expression, influencing body plan formation and organ development in animals, fungi, and plants. This domain functions as a transcription factor and is crucial for proper embryonic development. In animals, these genes often dictate where the limbs and other body segments will develop.
So what are the benefits of having protein domain information?
Protein domains are distinct functional and structural units in a protein. Information about domains can help to understand protein function, evolution, and interactions. The CDD helps in predicting the function of unknown proteins, offering a window into their potential roles in cellular processes.
This can be particularly useful for proteins that are not well annotated, or for proteins that are not well conserved across species. For example, if you have a protein that is only found in a single species, you may not be able to find any similar sequences in a BLAST search. But if that protein contains a conserved domain, you can still identify that domain, and thus get some information about the function of the protein.
CDD annotation can thus be used to:
- Get a quick summary of the likely functions of a protein.
- Identify the likely function of a protein that has no or only weak BLAST hits.
- Obtain protein annotations for large numbers of genes… which can help to name proteins in the predicted proteome of a newly sequenced species.
Multiple domains for multiple functions?
Complex proteins often have multiple domains. Each domain can have a distinct structure and function, allowing the protein to perform multiple roles or interact with different molecules within the cell.
- Functional Diversity: Different domains can confer different functions to the protein, such as binding to DNA, other proteins, or small molecules, catalyzing biochemical reactions, or signaling.
- Evolutionary Advantage: Multi-domain proteins can arise through gene fusion events, where two or more genes or parts of genes combine. This can lead to new functionalities and is a significant driver of evolutionary innovation.
- Regulatory Complexity: The presence of multiple domains can also allow for complex regulation of a protein’s activity, as different domains can be independently regulated.
How do I try SequenceServer’s domain identification?
Conserved domain information is immediately available to all SequenceServer Cloud users. To create a secure SequenceServer Cloud instance for yourself or your team, sign up today.
Happy BLASTing



