Visualizing and interpreting BLAST alignments
When running BLAST in the command-line, you end up with a large amount of text. If you have one query with one HSP in a single hit sequence of your BLAST database, interpretation is easy.
However, things are more complicated if:
- you have many queries,
- you have multiple HSPs per hit,
- you have multiple hits,
- you have a combination of the above.
It’s particularly confusing when multiple aligning segments (HSPs) aren’t in chromosomal order. But why should they be? Aligning segments are ordered by biological similarity (e-value). This is great for some contexts, but not necessarily, say when you are trying to understand how many times your query gene is present in the target genome.
It quickly becomes a mess trying to integrate large amounts of text and numbers by keeping track of coordinates on the hit sequences, coordinates on the query sequence, and reading frames and DNA strands…
Speaking from experience, the easiest can be to draw a summary picture on a piece of paper. But it’s 2021 - computers should be able to automate that!
A picture is worth 1000 words
Visualizations to make BLAST result interpretation easier exist thanks to many efforts, including those at NCBI, those we made for GeneValidator, the work of Nikos Darzentas with Circoletto and the work of Jeff Wintersinger & James Wasmuth with Kablammo.
We took inspiration from those efforts so that SequenceServer could better help biologists interpret BLAST results.
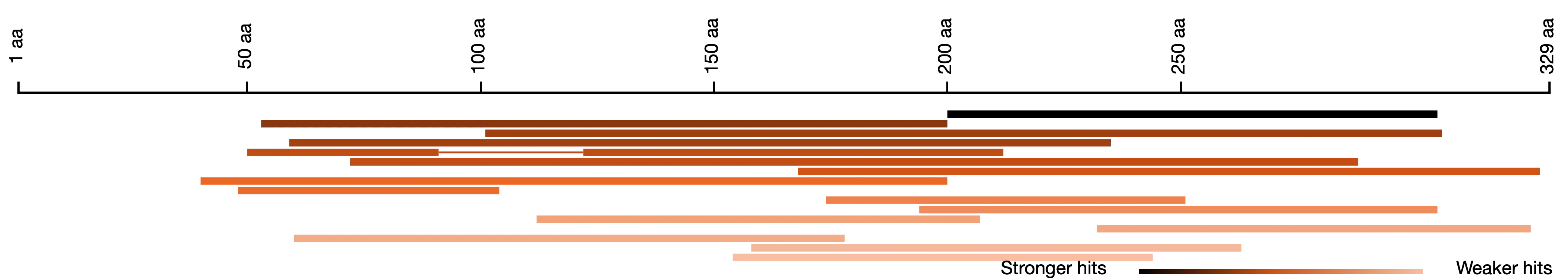
SequenceServer’s overview of how hit sequences align to the BLAST search query
Everyone who has used NCBI’s BLAST will have seen the overview of how hits align to the query sequence. SequenceServer provides a similar overview. A key difference is that in our visualization, darker segments means stronger e-value. This can be more intuitive to understand than the somewhat psychedelic colors that NCBI uses by default.
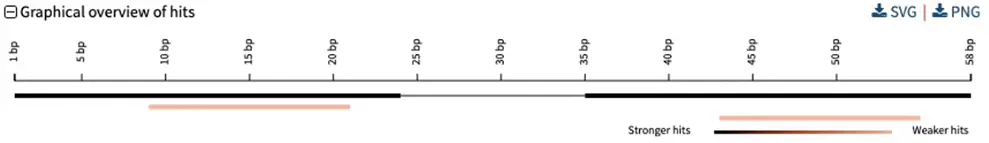
For specific types of analyses, e.g., checking PCR primer specificity, you may want to further adjust your strategy, which could give this type of visualization:
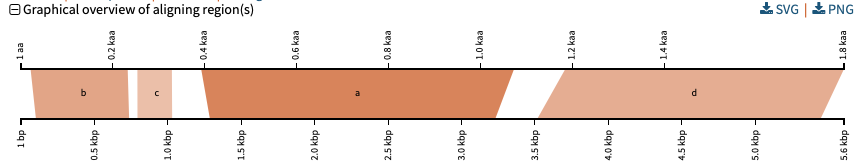
Pairwise alignments of BLAST query and hit sequences showing each HSP
For each hit sequence (horizontal at the bottom), we show how each aligning segment (HSP or High Scoring Pair) aligns to the query sequence (horizontal at the top). Darker alignment means stronger similarity. This visualization is derived from James Wasmuth’s work with Kablammo.
Comparing the lengths of BLAST queries and the lengths of hits
For each query sequence, we show a histogram of lengths of aligning sequences. Here, the query sequence is more than 1200 amino-acids long, while most hit sequences from the database are much shorter. Most of the hit sequences are 450 to 650 amino-acids long. The hit sequences with the highest BLAST similarity to the query sequence have the darkest color; their lengths cluster around 550-650 amino-acids long.
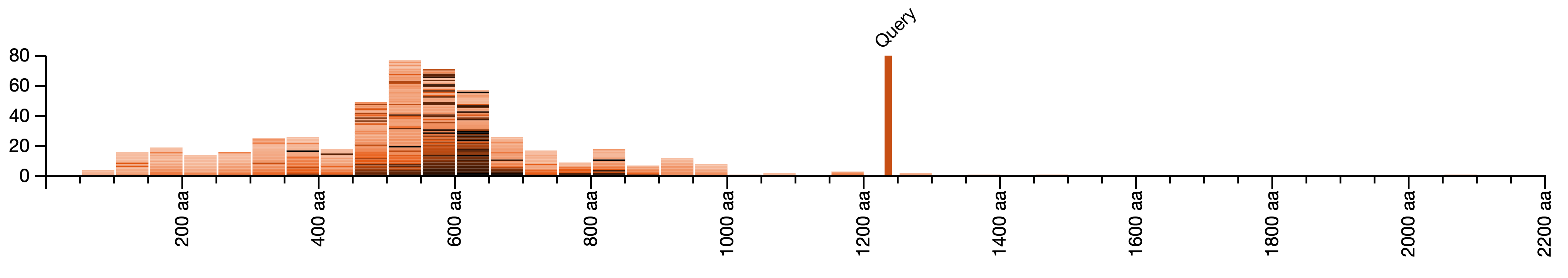
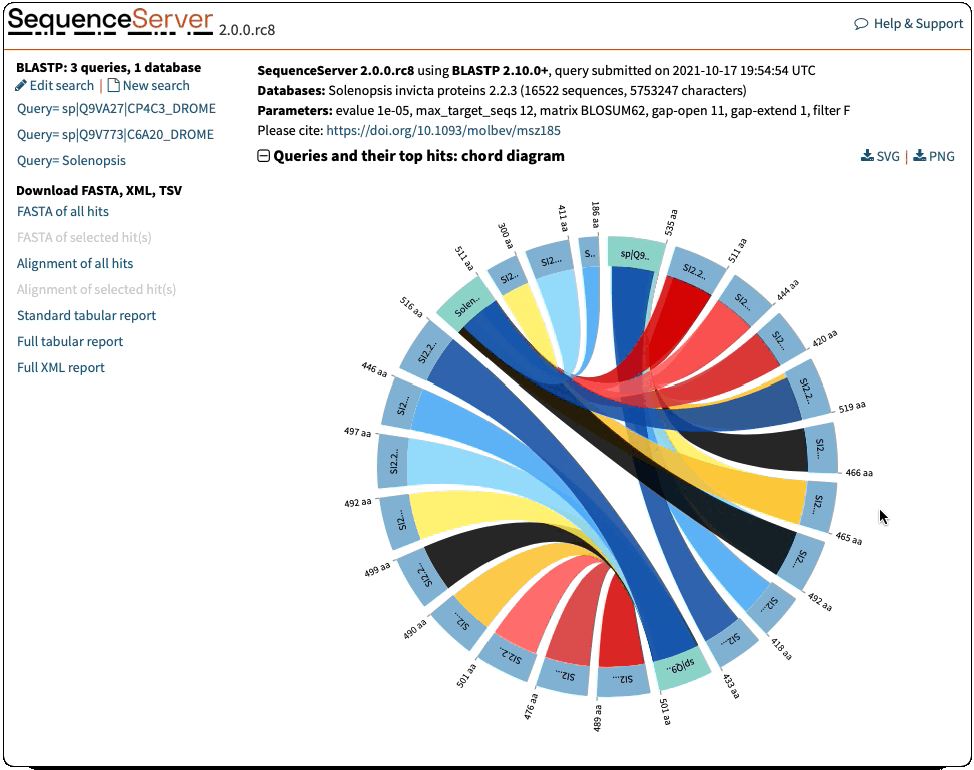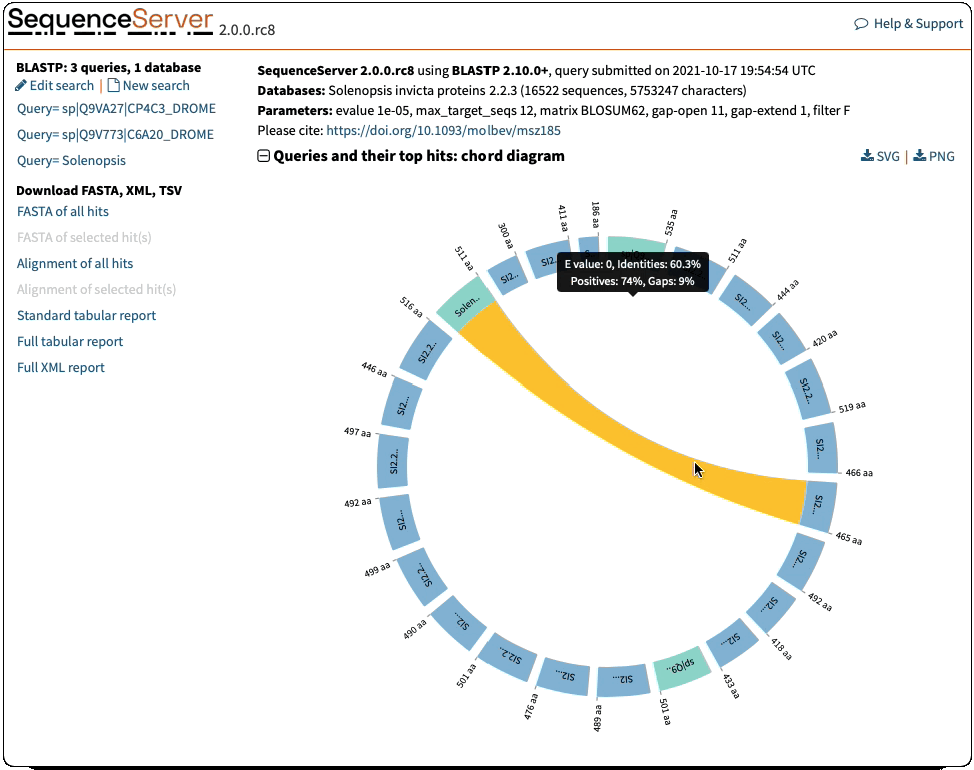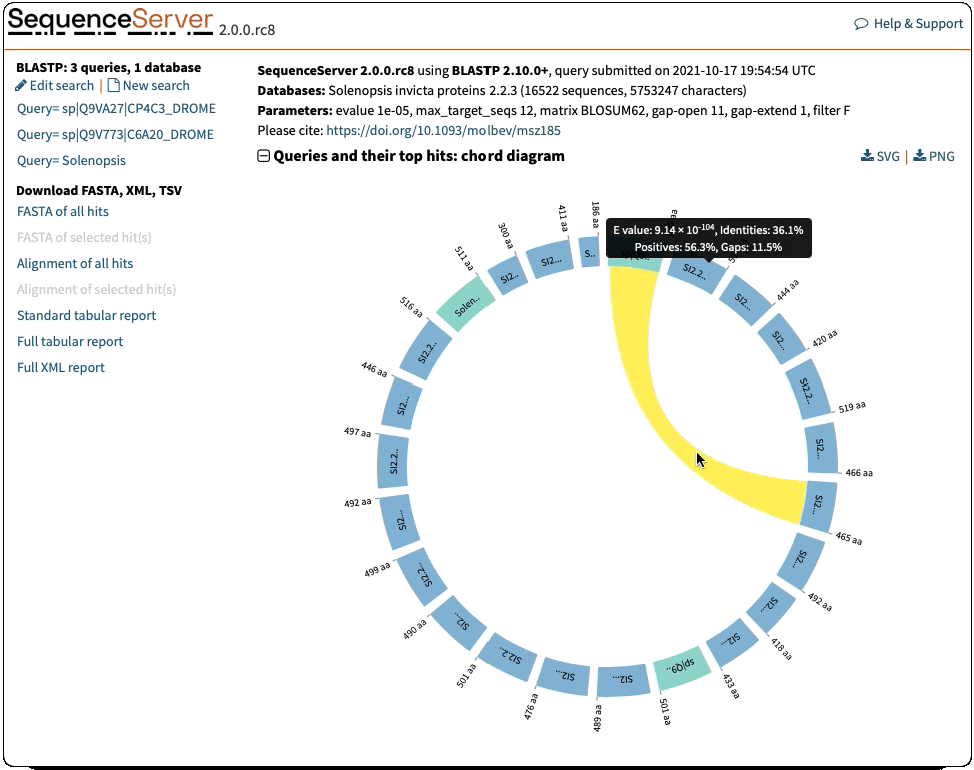
Circos/Circoletto-like overview of segments (HSPs) aligning between BLAST query and hit sequences
Here, three query sequences (in gray) show high similarity to a much larger number of query sequences.
Installing SequenceServer
You can readily perform BLAST analyses using SequenceServer, or use SequenceServer to visualise BLAST XML output. Install it yourself, or use our cloud service to perform and interpret BLAST analyses.
By leveraging cloud computing and publication-ready graphics, SequenceServer Cloud makes it easy to perform sequence search results and to interpret them .
