Examples of SequenceServer Usage
Check out the wide range of SequenceServer uses and applications
Exploring pea plant adaptations to low-sulfur environments with the help of SequenceServer!
2024-08-20
🌱Peas are plant protein powerhouses (phew, that’s a lot of P’s!), but quality relies on sulfur availability. Bachelet et al., with the help of SequenceServer, explore how pea compensates for low-sulfur environments using sulfate transport systems. Huge congrats to the authors! Check out the paper!

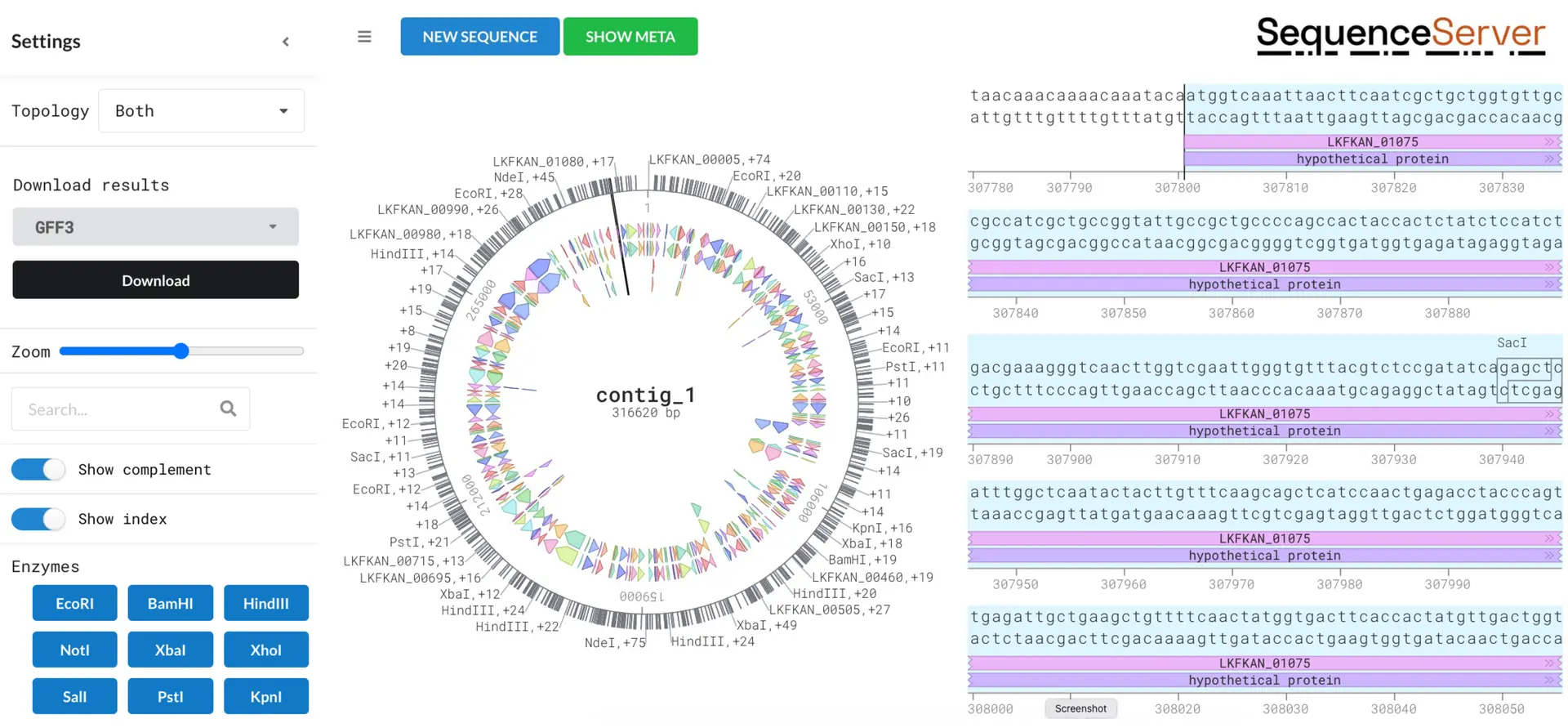
Genome annotation with SequenceServer cloud's DNA Visualization tool!
2024-08-13
Whether you’re new to genome annotation or a seasoned pro, SequenceServer Cloud’s DNA visualizer uses Bakta (Schwengers et al., 2021) to streamline the process to a simple copy, paste, & click! 🧬 Analyze BLAST hits, plasmids, & whole genome features with informative visualizations! Want to find out more? Check out our blog post for a detailed look at how this tool can enhance your research.
SequenceServer helps to uncover the mechanisms behind sex-determination in butterflies.
2024-07-16
We love celebrating the incredible work of researchers worldwide, & seeing SequenceServer’s growing impact! 🌟 One perfect example is van’t Hof et al., 2024, reporting a previously undescribed sex-determination mechanism in Lepidoptera! 🦋 Huge congrats to the authors!

SequenceServer helps to make it easier to develop pest resistant crops
2024-06-25
Always great to see SequenceServer in action! This time helping researchers identify the genes in barley 🌾 that defend against root-lesion nematodes! These pesky parasites infest roots, lay eggs, & cause significant crop & economic damage. That’s why selecting for resistance could be so valuable!

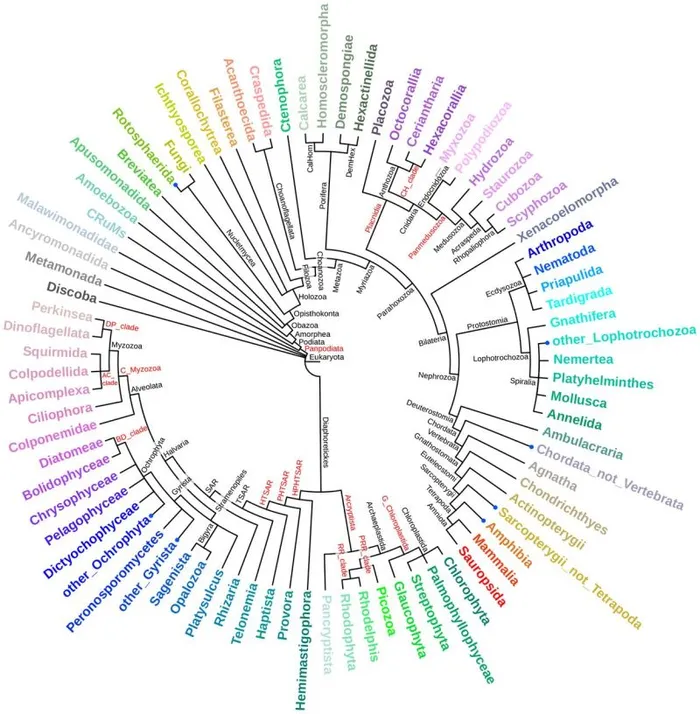
LukProt database powered by SequenceServer
2024-05-27
Discover LukProt - an extensive sequence database by Sobala et al, 2024, with advanced BLAST server capabilities powered by SequenceServer! 🧬🔍 It is a phenomenal resource for studying early animal evolution and a major win for data accessibility 🐶 Check it out for yourself.